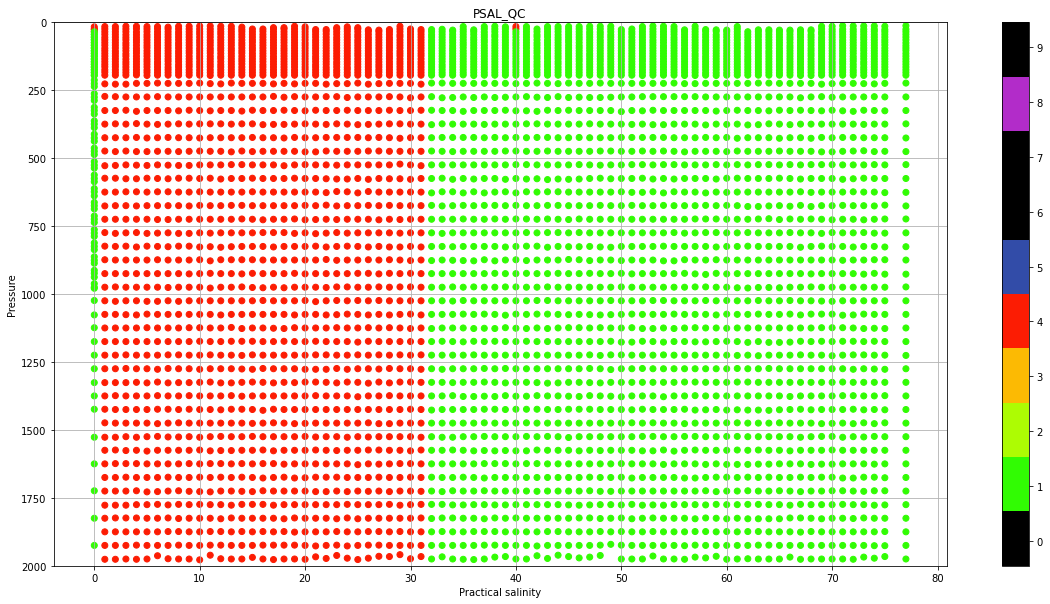
Real Time quality control data#
Real-Time quality control is a set of automatic procedures that are performed at the National Data Acquisition Centers (DACs) to carry out the first quality control of the data. There are a total of 19 tests that aim, to say, easy to identify anomalies in the data. The subtle anomalies, that need a lot of expertise and time to discern between sensor malfunctioning and natural variability, are left for the Delayed-Mode quality control.
The results of the Real-Time tests are summarized in what is called the quality control flags. Quality control flags are an essential part of Argo.
Quality Control flags#
Each observation after the RT quality control has a QC flag associated, a number from 0 to 9, with the following meaning:
QCflag |
Meaning |
Real time description |
|---|---|---|
0 |
No QC performed |
No QC performed |
1 |
Good data |
All real time QC tests passed |
2 |
Probably good data |
Probably good |
3 |
Bad data that are potentially correctable |
Test 15 or Test 16 or Test 17 failed and all other real-time QC tests passed. These data are not to be used without scientific correction. A flag ‘3’ may be assigned by an operator during additional visual QC for bad |
4 |
Bad data |
Data have failed one or more of the real-time QC tests, excluding Test 16. A flag ‘4’ may be assigned by an operator during additional visual QC for bad data that are not correctable. |
5 |
Value changed |
Value changed |
6 |
Not currently used |
Not currently used |
7 |
Not currently used |
Not currently used |
8 |
Estimated |
Estimated value (interpolated, extrapolated or other estimation) |
9 |
Missing value |
Missing value |
First, let’s see how this information is stored in the NetCDF files
import numpy as np
import netCDF4
import xarray as xr
import matplotlib as mpl
import matplotlib.cm as cm
from matplotlib import pyplot as plt
%matplotlib inline
Before accesing the data, let’s create some usefull colormaps and colorbar makers to help us to understand the QC flags
qcmap = mpl.colors.ListedColormap(['#000000',
'#31FC03',
'#ADFC03',
'#FCBA03',
'#FC1C03',
'#324CA8',
'#000000',
'#000000',
'#B22CC9',
'#000000'])
def colorbar_qc(cmap, **kwargs):
"""Adjust colorbar ticks with discrete colors for QC flags"""
ncolors = 10
mappable = cm.ScalarMappable(cmap=cmap)
mappable.set_array([])
mappable.set_clim(-0.5, ncolors+0.5)
colorbar = plt.colorbar(mappable, **kwargs)
colorbar.set_ticks(np.linspace(0, ncolors, ncolors))
colorbar.set_ticklabels(range(ncolors))
return colorbar
## QC flags for data accessed by date open the daily data set from the 11th november 2019
dayADS = xr.open_dataset('../../Data/atlantic_ocean/2020/11/20201111_prof.nc')
dayADS
<xarray.Dataset>
Dimensions: (N_PROF: 188, N_PARAM: 3, N_LEVELS: 1331, N_CALIB: 3, N_HISTORY: 0)
Dimensions without coordinates: N_PROF, N_PARAM, N_LEVELS, N_CALIB, N_HISTORY
Data variables: (12/64)
DATA_TYPE object b'Argo profile '
FORMAT_VERSION object b'3.1 '
HANDBOOK_VERSION object b'1.2 '
REFERENCE_DATE_TIME object b'19500101000000'
DATE_CREATION object b'20201111082709'
DATE_UPDATE object b'20211211022000'
... ...
HISTORY_ACTION (N_HISTORY, N_PROF) object
HISTORY_PARAMETER (N_HISTORY, N_PROF) object
HISTORY_START_PRES (N_HISTORY, N_PROF) float32
HISTORY_STOP_PRES (N_HISTORY, N_PROF) float32
HISTORY_PREVIOUS_VALUE (N_HISTORY, N_PROF) float32
HISTORY_QCTEST (N_HISTORY, N_PROF) object
Attributes:
title: Argo float vertical profile
institution: FR GDAC
source: Argo float
history: 2021-12-11T02:20:00Z creation
references: http://www.argodatamgt.org/Documentation
user_manual_version: 3.1
Conventions: Argo-3.1 CF-1.6
featureType: trajectoryProfile- N_PROF: 188
- N_PARAM: 3
- N_LEVELS: 1331
- N_CALIB: 3
- N_HISTORY: 0
- DATA_TYPE()object...
- long_name :
- Data type
- conventions :
- Argo reference table 1
array(b'Argo profile ', dtype=object)
- FORMAT_VERSION()object...
- long_name :
- File format version
array(b'3.1 ', dtype=object)
- HANDBOOK_VERSION()object...
- long_name :
- Data handbook version
array(b'1.2 ', dtype=object)
- REFERENCE_DATE_TIME()object...
- long_name :
- Date of reference for Julian days
- conventions :
- YYYYMMDDHHMISS
array(b'19500101000000', dtype=object)
- DATE_CREATION()object...
- long_name :
- Date of file creation
- conventions :
- YYYYMMDDHHMISS
array(b'20201111082709', dtype=object)
- DATE_UPDATE()object...
- long_name :
- Date of update of this file
- conventions :
- YYYYMMDDHHMISS
array(b'20211211022000', dtype=object)
- PLATFORM_NUMBER(N_PROF)object...
- long_name :
- Float unique identifier
- conventions :
- WMO float identifier : A9IIIII
array([b'3901238 ', b'4903277 ', b'6903030 ', b'5904669 ', b'3901819 ', b'5905132 ', b'5902338 ', b'6902976 ', b'6903698 ', b'6903032 ', b'4902909 ', b'7900506 ', b'6903034 ', b'6903788 ', b'4903035 ', b'4902920 ', b'6903270 ', b'6902655 ', b'3902107 ', b'3901938 ', b'1901814 ', b'6901986 ', b'1902070 ', b'3901969 ', b'6901985 ', b'6901144 ', b'4903259 ', b'3902399 ', b'3901043 ', b'6903268 ', b'6903788 ', b'4902117 ', b'3901312 ', b'4903228 ', b'4903261 ', b'5905380 ', b'4903245 ', b'6902762 ', b'4902348 ', b'6902834 ', b'3901971 ', b'4902354 ', b'4902323 ', b'3902148 ', b'6903788 ', b'3902121 ', b'6901935 ', b'3901870 ', b'6901934 ', b'4902116 ', b'3902238 ', b'3901551 ', b'3902121 ', b'5904847 ', b'6901194 ', b'5905383 ', b'4903260 ', b'1902228 ', b'4901628 ', b'3901822 ', b'6903016 ', b'6902727 ', b'6903788 ', b'3902237 ', b'3901850 ', b'6902851 ', b'6902855 ', b'6902854 ', b'6902852 ', b'6902929 ', b'7900559 ', b'3901869 ', b'3901859 ', b'6902899 ', b'3901878 ', b'3901857 ', b'3901838 ', b'3902168 ', b'4902912 ', b'6903550 ', b'3901111 ', b'6902818 ', b'5906005 ', b'4902921 ', b'3902106 ', b'6901774 ', b'3902137 ', b'6903567 ', b'6903705 ', b'3902106 ', b'4903240 ', b'4903276 ', b'3902207 ', b'3901541 ', b'6902898 ', b'3902137 ', b'5905148 ', b'6903788 ', b'6901932 ', b'5905381 ', b'7900207 ', b'3901823 ', b'4902350 ', b'3901658 ', b'7900563 ', b'6903554 ', b'4903225 ', b'3901605 ', b'4902424 ', b'1902063 ', b'4901699 ', b'6903552 ', b'4903237 ', b'4902916 ', b'6901925 ', b'3902109 ', b'6902974 ', b'6902686 ', b'6901604 ', b'6901763 ', b'4902456 ', b'3901633 ', b'3902110 ', b'6902837 ', b'3901526 ', b'3901540 ', b'3901644 ', b'1901712 ', b'3902109 ', b'6903558 ', b'3902208 ', b'6902944 ', b'6903266 ', b'6901281 ', b'6903788 ', b'3902110 ', b'3901977 ', b'6903786 ', b'3901867 ', b'7900596 ', b'6900893 ', b'6900892 ', b'4902455 ', b'6903255 ', b'7900516 ', b'7900534 ', b'6901266 ', b'3901675 ', b'7900542 ', b'6902870 ', b'3901673 ', b'6901254 ', b'6902873 ', b'7900524 ', b'6901255 ', b'5906215 ', b'6902875 ', b'4902523 ', b'3901937 ', b'3901622 ', b'4902524 ', b'4902505 ', b'5905076 ', b'4902509 ', b'6901278 ', b'4902501 ', b'6901208 ', b'1901819 ', b'4902510 ', b'4901702 ', b'4902471 ', b'3901601 ', b'3901931 ', b'1901822 ', b'6903723 ', b'6903788 ', b'3901226 ', b'4902345 ', b'3901983 ', b'6902964 ', b'6903273 ', b'6902965 ', b'4901625 ', b'4903236 ', b'5903661 ', b'4902346 ', b'6901726 ', b'6903788 '], dtype=object) - PROJECT_NAME(N_PROF)object...
- long_name :
- Name of the project
array([b'US ARGO PROJECT ', b'US ARGO PROJECT ', b'OVIDE ', b'UW, Argo ', b'US ARGO PROJECT ', b'UW, SOCCOM, Argo equivalent ', b'Argo ', b'OVIDE ', b'ARGO-FINLAND ', b'OVIDE ', b'US ARGO PROJECT ', b'ARGO-BSH ', b'OVIDE ', b'Argo Italy ', b'US ARGO PROJECT ', b'US ARGO PROJECT ', b'ARGO Italy ', b'Coriolis ', b'ARGO POLAND ', b'MOCCA-EU ', b'US ARGO PROJECT ', b'Argo Netherlands ', b'US ARGO PROJECT ', b'MOCCA-NETH ', b'Argo Netherlands ', b'Argo UK ', b'US ARGO PROJECT ', b'Argo UK ', b'US ARGO PROJECT ', b'Argo-Italy ', b'Argo Italy ', b'US ARGO PROJECT ', b'US ARGO PROJECT ', b'US ARGO PROJECT ', b'US ARGO PROJECT ', b'UW, Argo ', b'US ARGO PROJECT ', b'PIRATA ', b'US ARGO PROJECT ', b'CORIOLIS ', b'MOCCA-EU ', b'US ARGO PROJECT ', b'US ARGO PROJECT ', b'US ARGO PROJECT ', b'Argo Italy ', b'AtlantOS ', b'ARGO_IRELAND ', b'MOCCA-EU ', b'Argo Ireland ', b'US ARGO PROJECT ', b'US ARGO PROJECT ', b'Argo UK ', b'AtlantOS ', b'UW, SOCCOM, Argo equivalent ', b'Argo UK ', b'UW, Argo ', b'US ARGO PROJECT ', b'US ARGO PROJECT ', b'US ARGO PROJECT ', b'US ARGO PROJECT ', b'CORIOLIS ', b'CORIOLIS ', b'Argo Italy ', b'US ARGO PROJECT ', b'MOCCA-POLAND ', b'GMMC ARGOMEX ', b'GMMC ARGOMEX ', b'GMMC ARGOMEX ', b'GMMC ARGOMEX ', b'PIRATA ', b'Argo GERMANY ', b'MOCCA-EU ', b'MOCCA-EU ', b'Euro-Argo RISE ', b'MOCCA-NETH ', b'MOCCA-EU ', b'MOCCA-GER ', b'US ARGO PROJECT ', b'US ARGO PROJECT ', b'Norway-BGC-Argo ', b'US ARGO PROJECT ', b'GMMC OVIDE ', b'UW, SOCCOM, Argo equivalent ', b'US ARGO PROJECT ', b'ARGO POLAND ', b'NAOS ', b'MOCCA-EU ', b'Argo-Norway ', b'Argo-Finland ', b'ARGO POLAND ', b'US ARGO PROJECT ', b'US ARGO PROJECT ', b'US ARGO PROJECT ', b'Argo UK ', b'NAOS ', b'MOCCA-EU ', b'UW, Argo ', b'Argo Italy ', b'Argo Ireland ', b'UW, Argo ', b'US ARGO PROJECT ', b'US ARGO PROJECT ', b'US ARGO PROJECT ', b'ARGO-BSH ', b'Argo GERMANY ', b'ARGO Norway ', b'US ARGO PROJECT ', b'ARGO-BSH ', b'Argo Canada ', b'US ARGO PROJECT ', b'US ARGO PROJECT ', b'Argo-Norway ', b'US ARGO PROJECT ', b'US ARGO PROJECT ', b'Argo Ireland ', b'Euro-Argo RISE ', b'OVIDE ', b'RREX ', b'OVIDE ', b'GMMC OVIDE ', b'Argo Canada ', b'ARGO-BSH ', b'ARGO POLAND ', b'CORIOLIS ', b'Argo UK ', b'Argo UK ', b'ARGO-BSH ', b'US ARGO PROJECT ', b'Euro-Argo RISE ', b'Norway-BGC-Argo ', b'US ARGO PROJECT ', b'PERLE ', b'Argo Italy,CALYPSO 2019 ', b'ARGO SPAIN ', b'Argo Italy ', b'ARGO POLAND ', b'ARGO Italy ', b'Argo Italy ', b'MOCCA-EU ', b'ARGO Bulgary ', b'ARGO-BSH ', b'ARGO-BSH ', b'Argo Canada ', b'ARGO Italy ', b'ARGO-BSH ', b'ARGO-BSH ', b'ARGO SPAIN ', b'ARGO-BSH ', b'ARGO-BSH ', b'GMMC PERLE ', b'ARGO-BSH ', b'ARGO SPAIN ', b'GMMC PERLE ', b'ARGO-BSH ', b'ARGO SPAIN ', b'UW, SOCCOM, Argo equivalent ', b'GMMC PERLE ', b'Argo Canada ', b'MOCCA-EU ', b'ARGO-BSH ', b'Argo Canada ', b'Argo Canada ', b'UW, SOCCOM, Argo equivalent ', b'Argo Canada ', b'Euro-Argo RISE ', b'Argo Canada ', b'Argo UK ', b'US ARGO PROJECT ', b'Argo Canada ', b'US ARGO PROJECT ', b'Argo Canada ', b'ARGO-BSH ', b'MOCCA-EU ', b'US ARGO PROJECT ', b'Argo UK ', b'Argo Italy ', b'US ARGO PROJECT ', b'US ARGO PROJECT ', b'MOCCA-EU ', b'CORIOLIS ', b'ARGO Norway ', b'MOOSE ', b'US ARGO PROJECT ', b'US ARGO PROJECT ', b'Argo Australia ', b'US ARGO PROJECT ', b'RREX ASFAR ', b'Argo Italy '], dtype=object) - PI_NAME(N_PROF)object...
- long_name :
- Name of the principal investigator
array([b'BRECK OWENS, STEVEN JAYNE, P.E. ROBBINS ', b'WHOI: WIJFFELS, JAYNE, ROBBINS ', b'Damien DESBRUYERES ', b'STEPHEN RISER ', b'BRECK OWENS, STEVEN JAYNE, P.E. ROBBINS ', b'STEPHEN RISER, KENNETH JOHNSON ', b'DEAN ROEMMICH ', b'Damien DESBRUYERES ', b'Laura Tuomi ', b'Damien DESBRUYERES ', b'BRECK OWENS, STEVEN JAYNE, P.E. ROBBINS ', b'Birgit KLEIN ', b'Damien DESBRUYERES ', b'Pierre-Marie Poulain ', b'BRECK OWENS, STEVEN JAYNE, P.E. ROBBINS ', b'BRECK OWENS, STEVEN JAYNE, P.E. ROBBINS ', b'Pierre-Marie Poulain ', b'Christine COATANOAN ', b'Waldemar Walczowski ', b'Romain Cancouet ', b'BRECK OWENS, STEVEN JAYNE, P.E. ROBBINS ', b'Andreas Sterl ', b'BRECK OWENS, STEVEN JAYNE, P.E. ROBBINS ', b'Andreas Sterl ', b'Andreas Sterl ', b'Jon Turton ', b'AMY BOWER, STEVEN JAYNE, HEATHER FUREY ', b'Jon Turton ', b'BRECK OWENS, STEVE JAYNE, P.E. ROBBINS ', b'Pierre-Marie Poulain ', b'Pierre-Marie Poulain ', b'BRECK OWENS, STEVEN JAYNE, P.E. ROBBINS ', b'GREGORY C. JOHNSON ', b'WHOI: WIJFFELS, JAYNE, ROBBINS ', b'WHOI: WIJFFELS, JAYNE, ROBBINS ', b'STEPHEN RISER, ', b'WIJFFELS, JAYNCE, ROBBINS ', b'Bernard BOURLES ', b'BRECK OWENS, STEVEN JAYNE, P.E. ROBBINS ', b'Christine COATANOAN ', b'Romain Cancouet ', b'BRECK OWENS, STEVEN JAYNE, P.E. ROBBINS ', b'DEAN ROEMMICH ', b'GREGORY C. JOHNSON ', b'Pierre-Marie Poulain ', b'Herve CLAUSTRE ', b"Conall O'Malley ", b'Peter Brandt ', b"Conall O'Malley ", b'BRECK OWENS, STEVEN JAYNE, P.E. ROBBINS ', b'WHOI: WIJFFELS, JAYNE, ROBBINS ', b'Jon Turton ', b'Herve CLAUSTRE ', b'STEPHEN RISER, KENNETH JOHNSON ', b'Jon Turton ', b'STEPHEN RISER, ', b'WHOI: WIJFFELS, JAYNE, ROBBINS ', b'WHOI: WIJFFELS, JAYNE, ROBBINS ', b'BRECK OWENS, STEVEN JAYNE, P.E. ROBBINS ', b'BRECK OWENS, STEVEN JAYNE, P.E. ROBBINS ', b'Franck DUMAS ', b'Marcel BABIN ', b'Pierre-Marie Poulain ', b'WIJFFELS, JAYNE, ROBBINS ', b'Waldemar Walczowski ', b"Fabrizio D'ORTENZIO ", b"Fabrizio D'ORTENZIO ", b"Fabrizio D'ORTENZIO ", b"Fabrizio D'ORTENZIO ", b'Bernard BOURLES ', b'Oliver Zielinski ', b'Peter Brandt ', b'Romain Cancouet ', b"Fabrizio D'ORTENZIO ", b'Andreas Sterl ', b'Romain Cancouet ', b'Birgit Klein ', b'WIJFFELS, JAYNE, ROBBINS ', b'BRECK OWENS, STEVEN JAYNE, P.E. ROBBINS ', b'Kjell Arne Mork ', b'BRECK OWENS, STEVEN JAYNE, P.E. ROBBINS ', b'Virginie THIERRY ', b'STEPHEN RISER, KENNETH JOHNSON ', b'BRECK OWENS, STEVEN JAYNE, P.E. ROBBINS ', b'Waldemar Walczowski ', b"Fabrizio D'ORTENZIO ", b'Romain Cancouet ', b'Kjell Arne Mork ', b'J. Haapala ', b'Waldemar Walczowski ', b'AMY BOWER, STEVEN JAYNE, HEATHER FUREY ', b'WHOI: WIJFFELS, JAYNE, ROBBINS ', b'GREGORY C. JOHNSON ', b'Brian King ', b"Fabrizio D'ORTENZIO ", b'Romain Cancouet ', b'STEPHEN RISER ', b'Pierre-Marie Poulain ', b"Diarmuid O'Conchubhair ", b'STEPHEN RISER, ', b'DEAN ROEMMICH ', b'BRECK OWENS, STEVEN JAYNE, P.E. ROBBINS ', b'BRECK OWENS, STEVEN JAYNE, P.E. ROBBINS ', b'Birgit Klein ', b'Oliver Zielinski ', b'Kjell Arne Mork ', b'WHOI: WIJFFELS, JAYNE, ROBBINS ', b'Birgit KLEIN ', b'Blair Greenan ', b'BRECK OWENS, STEVEN JAYNE, P.E. ROBBINS ', b'BRECK OWENS, STEVEN JAYNE, P.E. ROBBINS ', b'Kjell Arne Mork ', b'AMY BOWER, STEVEN JAYNE, HEATHER FUREY ', b'BRECK OWENS, STEVEN JAYNE, P.E. ROBBINS ', b"Diarmuid O'Conchubhair ", b'Waldemar Walczowski ', b'Damien DESBRUYERES ', b'Virginie THIERRY ', b'Damien DESBRUYERES ', b'Virginie THIERRY ', b'Blair Greenan ', b'Birgit Klein ', b'Waldemar Walczowski ', b'Christine COATANOAN ', b'Jon Turton ', b'Brian King ', b'Birgit Klein ', b'BRECK OWENS, STEVE JAYNE, P.E. ROBBINS ', b'Waldemar Walczowski ', b'Kjell Arne Mork ', b'GREGORY C. JOHNSON ', b'Franck DUMAS ', b'Pierre-Marie Poulain ', b'Inmaculada Ruiz-Parrado ', b'Pierre-Marie Poulain ', b'Waldemar Walczowski ', b'Pierre-Marie Poulain ', b'Pierre-Marie Poulain ', b'Peter Brandt ', b'Violeta SLABAKOVA ', b'Birgit Klein ', b'Birgit Klein ', b'Blair Greenan ', b'Pierre-Marie Poulain ', b'Birgit Klein ', b'Birgit Klein ', b'Pedro Velez ', b'Birgit Klein ', b'Dr. Birgit Klein ', b'Laurent COPPOLA ', b'Birgit Klein ', b'Pedro Velez ', b'Laurent COPPOLA ', b'Birgit Klein ', b'Pedro Velez ', b'STEPHEN RISER, KENNETH JOHNSON ', b'Laurent COPPOLA ', b'Blair Greenan ', b'Sabrina Speich ', b'Birgit Klein ', b'Blair Greenan ', b'Blair Greenan ', b'STEPHEN RISER, KENNETH JOHNSON ', b'Blair Greenan ', b'Inmaculada Ruiz-Parrado ', b'Blair Greenan ', b'Jon Turton ', b'BRECK OWENS, STEVEN JAYNE, P.E. ROBBINS ', b'Blair Greenan ', b'BRECK OWENS, STEVEN JAYNE, P.E. ROBBINS ', b'Blair Greenan ', b'Birgit KLEIN ', b'Sabrina Speich ', b'BRECK OWENS, STEVEN JAYNE, P.E. ROBBINS ', b'Jon Turton ', b'Pierre-Marie Poulain ', b'BRECK OWENS, STEVEN JAYNE, P.E. ROBBINS ', b'BRECK OWENS, STEVEN JAYNE, P.E. ROBBINS ', b'Josep Lluis Pelegri ', b'Sabrina SPEICH ', b'Kjell Arne MORK ', b'Laurent COPPOLA ', b'BRECK OWENS, STEVEN JAYNE, P.E. ROBBINS ', b'AMY BOWER, STEVEN JAYNE, HEATHER FUREY ', b'Susan Wijffels ', b'BRECK OWENS, STEVEN JAYNE, P.E. ROBBINS ', b'Virginie THIERRY ', b'Pierre-Marie Poulain '], dtype=object) - STATION_PARAMETERS(N_PROF, N_PARAM)object...
- long_name :
- List of available parameters for the station
- conventions :
- Argo reference table 3
array([[b'PRES ', b'TEMP ', b'PSAL '], [b'PRES ', b'TEMP ', b'PSAL '], [b'PRES ', b'TEMP ', b'PSAL '], ..., [b'PRES ', b'TEMP ', b'PSAL '], [b'PRES ', b'TEMP ', b'PSAL '], [b'PRES ', b'TEMP ', b'PSAL ']], dtype=object) - CYCLE_NUMBER(N_PROF)float64...
- long_name :
- Float cycle number
- conventions :
- 0...N, 0 : launch cycle (if exists), 1 : first complete cycle
array([140., 10., 9., 180., 248., 113., 281., 9., 272., 9., 130., 138., 9., 110., 117., 116., 86., 181., 93., 114., 148., 39., 114., 95., 38., 293., 85., 38., 260., 78., 109., 163., 54., 29., 29., 103., 47., 133., 160., 80., 85., 241., 101., 49., 108., 266., 7., 147., 8., 163., 41., 76., 265., 69., 157., 103., 38., 40., 247., 130., 27., 175., 107., 46., 161., 165., 165., 165., 165., 151., 168., 147., 154., 114., 131., 156., 155., 48., 253., 110., 194., 87., 62., 116., 407., 146., 401., 37., 22., 406., 102., 10., 47., 141., 246., 400., 114., 106., 54., 103., 216., 248., 153., 103., 31., 120., 32., 152., 92., 147., 238., 4., 102., 250., 172., 238., 13., 121., 13., 88., 77., 114., 85., 68., 149., 141., 69., 307., 237., 54., 33., 240., 263., 2., 105., 84., 151., 12., 150., 35., 41., 40., 77., 63., 69., 53., 89., 89., 36., 128., 90., 79., 136., 54., 60., 31., 128., 4., 141., 118., 4., 11., 136., 11., 100., 4., 86., 165., 11., 239., 52., 152., 140., 148., 2., 104., 194., 159., 116., 109., 196., 13., 246., 103., 357., 162., 158., 103.]) - DIRECTION(N_PROF)object...
- long_name :
- Direction of the station profiles
- conventions :
- A: ascending profiles, D: descending profiles
array([b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'D', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'D', b'D', b'A', b'D', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'D', b'A', b'D', b'A', b'A', b'A', b'A', b'A', b'D', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'D', b'A', b'A', b'A', b'A', b'A', b'A', b'D', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A'], dtype=object) - DATA_CENTRE(N_PROF)object...
- long_name :
- Data centre in charge of float data processing
- conventions :
- Argo reference table 4
array([b'AO', b'AO', b'IF', b'AO', b'AO', b'AO', b'AO', b'IF', b'IF', b'IF', b'AO', b'IF', b'IF', b'IF', b'AO', b'AO', b'IF', b'IF', b'IF', b'BO', b'AO', b'IF', b'AO', b'BO', b'IF', b'BO', b'AO', b'BO', b'AO', b'IF', b'IF', b'AO', b'AO', b'AO', b'AO', b'AO', b'AO', b'IF', b'AO', b'IF', b'BO', b'AO', b'AO', b'AO', b'IF', b'IF', b'BO', b'IF', b'BO', b'AO', b'AO', b'BO', b'IF', b'AO', b'BO', b'AO', b'AO', b'AO', b'AO', b'AO', b'IF', b'IF', b'IF', b'AO', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'BO', b'IF', b'IF', b'AO', b'AO', b'IF', b'AO', b'IF', b'AO', b'AO', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'AO', b'AO', b'AO', b'BO', b'IF', b'IF', b'AO', b'IF', b'BO', b'AO', b'AO', b'AO', b'AO', b'IF', b'IF', b'IF', b'AO', b'IF', b'ME', b'AO', b'AO', b'IF', b'AO', b'AO', b'BO', b'IF', b'IF', b'IF', b'IF', b'IF', b'ME', b'IF', b'IF', b'IF', b'BO', b'BO', b'IF', b'AO', b'IF', b'IF', b'AO', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'ME', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'IF', b'AO', b'IF', b'ME', b'IF', b'IF', b'ME', b'ME', b'AO', b'ME', b'IF', b'ME', b'BO', b'AO', b'ME', b'AO', b'ME', b'IF', b'IF', b'AO', b'BO', b'IF', b'AO', b'AO', b'BO', b'IF', b'IF', b'IF', b'AO', b'AO', b'CS', b'AO', b'IF', b'IF'], dtype=object) - DC_REFERENCE(N_PROF)object...
- long_name :
- Station unique identifier in data centre
- conventions :
- Data centre convention
array([b'7104 ', b'8480 ', b' ', b'6179 ', b'7133 ', b'7073 ', b'5166 ', b' ', b' ', b' ', b'7111 ', b' ', b' ', b' ', b'7490 ', b'7472 ', b' ', b' ', b' ', b' ', b'6481 ', b' ', b'7477 ', b' ', b' ', b'137170 ', b'8047 ', b'137166 ', b'5136 ', b' ', b' ', b'6730 ', b'7940 ', b'8003 ', b'8458 ', b'7441 ', b'8020 ', b' ', b'6747 ', b' ', b' ', b'7109 ', b'6848 ', b'7943 ', b' ', b' ', b' ', b' ', b' ', b'6729 ', b'8030 ', b'137164 ', b' ', b'6704 ', b'137158 ', b'7444 ', b'8053 ', b'8034 ', b'5359 ', b'7136 ', b' ', b' ', b' ', b'8029 ', b' ', b' ', b' ', b' ', b' ', b' ', b' ', b' ', b' ', b' ', b' ', b' ', b' ', b'8025 ', b'7114 ', b' ', b'6440 ', b' ', b'7688 ', b'7473 ', b' ', b' ', b' ', b' ', b' ', b' ', b'8015 ', b'8479 ', b'7966 ', b'138577 ', b' ', b' ', b'7089 ', b' ', b' ', b'7442 ', b'5606_008321_216 ', b'7137 ', b'6749 ', b' ', b' ', b' ', b'8000 ', b' ', b'4902424_9931_TE ', b'7094 ', b'5491 ', b' ', b'8012 ', b'7130 ', b'137152 ', b' ', b' ', b' ', b' ', b' ', b'4902456_9930_TE ', b' ', b' ', b' ', b'137159 ', b'138569 ', b' ', b'5343 ', b' ', b' ', b'7975 ', b' ', b' ', b' ', b' ', b' ', b' ', b' ', b' ', b' ', b' ', b' ', b'4902455_9926_TE ', b' ', b' ', b' ', b' ', b' ', b' ', b' ', b' ', b' ', b' ', b' ', b' ', b'8359 ', b' ', b'4902523_9991_TE ', b' ', b' ', b'4902524_9991_TE ', b'4902505_9979_TE ', b'7017 ', b'4902509_9979_TE ', b' ', b'4902501_9991_TE ', b'137153 ', b'6726 ', b'4902510_9975_TE ', b'5494 ', b'4902471_9678_TE ', b' ', b' ', b'6753 ', b'137154 ', b' ', b'6451 ', b'6744 ', b' ', b' ', b' ', b' ', b'5356 ', b'8011 ', b'5903661/357 ', b'6745 ', b' ', b' '], dtype=object) - DATA_STATE_INDICATOR(N_PROF)object...
- long_name :
- Degree of processing the data have passed through
- conventions :
- Argo reference table 6
array([b'2C ', b'2B ', b'2B ', b'2C ', b'2C ', b'2C ', b'2B ', b'2B ', b'2B ', b'2B ', b'2C ', b'2C ', b'2B ', b'2B ', b'2B ', b'2C ', b'2B ', b'2C ', b'2C ', b'2B ', b'2B ', b'2C ', b'2C ', b'2C ', b'2C ', b'2B ', b'2B ', b'2B+ ', b'2B ', b'2B ', b'2B ', b'2B ', b'2C ', b'2B ', b'2B ', b'2C ', b'2B ', b'2C ', b'2B ', b'2C ', b'2B ', b'2B ', b'2B ', b'2C ', b'2B ', b'2C ', b'2B ', b'2C ', b'2B ', b'2B ', b'2B ', b'2C ', b'2C ', b'2C ', b'2B ', b'2C ', b'2B ', b'2B ', b'2B ', b'2C ', b'2B ', b'2B ', b'2B ', b'2C ', b'2C ', b'2B ', b'2B ', b'2B ', b'2B ', b'2C ', b'2C ', b'2C ', b'2C ', b'2B ', b'2B ', b'2C ', b'2C ', b'2B ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2B ', b'2B ', b'2B ', b'2B ', b'2B ', b'2B ', b'2B ', b'2C ', b'2B ', b'2C ', b'2C ', b'2B ', b'2B ', b'2C ', b'2B ', b'2B ', b'2C ', b'2C ', b'2C ', b'2B ', b'2C ', b'2B ', b'2B ', b'2B ', b'2C ', b'2B ', b'2C ', b'2C ', b'2B ', b'2C ', b'2C ', b'2B ', b'2B ', b'2B ', b'2C ', b'2B ', b'2C ', b'2B ', b'2C ', b'2B ', b'2C ', b'2C ', b'2C ', b'2C ', b'2B ', b'2B ', b'2C ', b'2B ', b'2B ', b'2B ', b'2B ', b'2B ', b'2B ', b'2B ', b'2B ', b'2C ', b'2B ', b'2C ', b'2C ', b'2B ', b'2B ', b'2C ', b'2C ', b'2C ', b'2C ', b'2C ', b'2B ', b'2C ', b'2B ', b'2B ', b'2C ', b'2C ', b'2C ', b'2B ', b'2B ', b'2B ', b'2C ', b'2B ', b'2B ', b'2C ', b'2B ', b'2B ', b'2C+ ', b'2B ', b'2C ', b'2B ', b'2B ', b'2B ', b'2C ', b'2B ', b'2B ', b'2B ', b'2B ', b'2C ', b'2B ', b'2B ', b'2B ', b'2C ', b'2B ', b'2B ', b'2C ', b'2C ', b'2B ', b'2B ', b'2B '], dtype=object) - DATA_MODE(N_PROF)object...
- long_name :
- Delayed mode or real time data
- conventions :
- R : real time; D : delayed mode; A : real time with adjustment
array([b'D', b'R', b'A', b'D', b'D', b'D', b'R', b'A', b'R', b'A', b'D', b'D', b'A', b'R', b'R', b'D', b'R', b'D', b'D', b'R', b'R', b'D', b'D', b'D', b'D', b'A', b'R', b'R', b'R', b'A', b'R', b'A', b'D', b'R', b'R', b'D', b'R', b'D', b'R', b'D', b'R', b'A', b'A', b'D', b'R', b'D', b'R', b'D', b'R', b'R', b'R', b'D', b'D', b'D', b'A', b'D', b'R', b'R', b'A', b'D', b'R', b'R', b'R', b'D', b'D', b'A', b'A', b'A', b'A', b'D', b'D', b'D', b'D', b'R', b'R', b'D', b'D', b'R', b'D', b'D', b'D', b'D', b'D', b'R', b'R', b'R', b'R', b'R', b'A', b'R', b'D', b'R', b'D', b'D', b'R', b'R', b'D', b'R', b'R', b'D', b'D', b'D', b'R', b'D', b'A', b'A', b'R', b'D', b'A', b'D', b'D', b'A', b'D', b'D', b'A', b'R', b'A', b'D', b'A', b'D', b'A', b'D', b'R', b'D', b'D', b'D', b'D', b'R', b'R', b'D', b'A', b'R', b'R', b'R', b'R', b'R', b'R', b'R', b'D', b'R', b'D', b'D', b'A', b'A', b'D', b'D', b'D', b'D', b'D', b'R', b'D', b'R', b'R', b'D', b'D', b'D', b'R', b'R', b'R', b'D', b'R', b'R', b'D', b'R', b'R', b'D', b'R', b'D', b'R', b'R', b'R', b'D', b'A', b'R', b'R', b'R', b'D', b'A', b'R', b'R', b'D', b'R', b'R', b'D', b'D', b'R', b'A', b'R'], dtype=object) - PLATFORM_TYPE(N_PROF)object...
- long_name :
- Type of float
- conventions :
- Argo reference table 23
array([b'S2A ', b'S2A ', b'ARVOR_D ', b'APEX ', b'S2A ', b'APEX ', b'SOLO_II ', b'ARVOR_D ', b'ARVOR ', b'ARVOR_D ', b'S2A ', b'APEX ', b'ARVOR_D ', b'ARVOR ', b'S2A ', b'S2A ', b'ARVOR ', b'ARVOR ', b'ARVOR ', b'ARVOR ', b'S2A ', b'ARVOR ', b'S2A ', b'ARVOR ', b'ARVOR ', b'APEX ', b'S2A ', b'APEX ', b'S2A ', b'ARVOR_D ', b'ARVOR ', b'S2A ', b'SOLO_D_MRV ', b'ALTO ', b'S2A ', b'APEX ', b'ALTO ', b'ARVOR ', b'S2A ', b'ARVOR ', b'ARVOR ', b'S2A ', b'SOLO_D ', b'SOLO_D_MRV ', b'ARVOR ', b'PROVOR_III ', b'ARVOR ', b'ARVOR ', b'ARVOR ', b'S2A ', b'S2A ', b'APEX ', b'PROVOR_III ', b'NAVIS_A ', b'APEX ', b'APEX ', b'S2A ', b'ALTO ', b'S2A ', b'S2A ', b'ARVOR ', b'ARVOR ', b'ARVOR ', b'S2A ', b'ARVOR ', b'ARVOR ', b'ARVOR ', b'ARVOR ', b'ARVOR ', b'ARVOR ', b'APEX ', b'ARVOR ', b'ARVOR ', b'PROVOR_III ', b'ARVOR ', b'ARVOR ', b'ARVOR ', b'S2A ', b'S2A ', b'PROVOR_III ', b'S2A ', b'ARVOR_D ', b'APEX ', b'S2A ', b'ARVOR ', b'PROVOR_III ', b'ARVOR ', b'APEX ', b'APEX ', b'ARVOR ', b'S2A ', b'S2A ', b'SOLO_D_MRV ', b'APEX ', b'PROVOR_III ', b'ARVOR ', b'APEX ', b'ARVOR ', b'ARVOR ', b'APEX ', b'SOLO_II ', b'S2A ', b'S2A ', b'ARVOR ', b'APEX ', b'APEX ', b'S2A ', b'APEX ', b'NOVA ', b'S2A ', b'S2A ', b'APEX ', b'S2A ', b'S2A ', b'APEX ', b'ARVOR ', b'ARVOR_D ', b'ARVOR ', b'ARVOR_D ', b'ARVOR_D ', b'NOVA ', b'ARVOR ', b'ARVOR ', b'ARVOR ', b'APEX ', b'APEX ', b'ARVOR ', b'S2A ', b'ARVOR ', b'ARVOR_D ', b'NAVIS_A ', b'ARVOR ', b'ARVOR ', b'ARVOR ', b'ARVOR ', b'ARVOR ', b'ARVOR ', b'ARVOR ', b'ARVOR ', b'ARVOR ', b'ARVOR ', b'ARVOR ', b'NOVA ', b'ARVOR ', b'ARVOR ', b'ARVOR ', b'ARVOR ', b'ARVOR ', b'ARVOR ', b'PROVOR ', b'ARVOR ', b'ARVOR ', b'PROVOR ', b'ARVOR ', b'ARVOR ', b'APEX ', b'PROVOR ', b'ARVOR ', b'ARVOR ', b'ARVOR ', b'ARVOR ', b'ARVOR ', b'APEX ', b'ARVOR ', b'ARVOR ', b'ARVOR ', b'APEX ', b'S2A ', b'ARVOR ', b'S2A ', b'ARVOR ', b'APEX ', b'ARVOR ', b'S2A ', b'APEX ', b'ARVOR ', b'S2A ', b'S2A ', b'ARVOR ', b'PROVOR ', b'APEX ', b'PROVOR ', b'S2A ', b'S2A ', b'APEX ', b'S2A ', b'ARVOR ', b'ARVOR '], dtype=object) - FLOAT_SERIAL_NO(N_PROF)object...
- long_name :
- Serial number of the float
array([b'7387 ', b'7538 ', b'AD2700-19FR003 ', b'7307 ', b'10088 ', b'8050 ', b'8190 ', b'AD2700-18FR015 ', b'AI2600-18FI001 ', b'AD2700-19FR005 ', b'7406 ', b'7956 ', b'AD2700-19FR007 ', b'AI2600-20EU004 ', b'7465 ', b'10091 ', b'AI2600-19EU004 ', b'OIN-15-AR-14 ', b'AI2600-18EU007 ', b'AI2600-16FR081 ', b'7330 ', b'AI2600-19EU017 ', b'7434 ', b'AI2600-16FR112 ', b'AI2600-19EU016 ', b'5012 ', b'7586 ', b'8476 ', b'7191 ', b'AD2700-18EU003 ', b'AI2600-20EU004 ', b'7359 ', b'12010 ', b'11027 ', b'7508 ', b'8279 ', b'11046 ', b'AR2000-16FR019 ', b'7419 ', b'AL2500-17FR005 ', b'AI2600-16FR114 ', b'7382 ', b'6026 ', b'12013 ', b'AI2600-20EU004 ', b'P41306-16FR102 ', b'AI2600-18EU032 ', b'AI2600-16FR033 ', b'AI2600-18EU030 ', b'7358 ', b'7562 ', b'8065 ', b'P41306-16FR102 ', b'0689 ', b'7577 ', b'8283 ', b'7585 ', b'11058 ', b'7205 ', b'10094 ', b'AI2600-19FR109 ', b'AI2600-16FR310 ', b'AI2600-20EU004 ', b'7580 ', b'AI2600-16FR013 ', b'AI2600-17FR008 ', b'AI2600-17FR012 ', b'AI2600-17FR011 ', b'AI2600-17FR009 ', b'AI2600-18FR020 ', b'8637 ', b'AI2600-16FR032 ', b'AI2600-16FR022 ', b'OIN13-S4-06 ', b'AI2600-16FR041 ', b'AI2600-16FR020 ', b'AI2600-16FR001 ', b'7587 ', b'7424 ', b'P43208-18NO002 ', b'7290 ', b'AD1726-16FR009 ', b'7971 ', b'10092 ', b'AI2632-18EU005 ', b'OIN-14SO-S4-11 ', b'AI2600-17EU013 ', b'8908 ', b'f8349 ', b'AI2632-18EU005 ', b'7568 ', b'7519 ', b'12036 ', b'8139 ', b'OIN-12-RA-S31-15 ', b'AI2600-17EU013 ', b'7688 ', b'AI2600-20EU004 ', b'AI2600-17EU04 ', b'8281 ', b'8321 ', b'10095 ', b'7369 ', b'AL2500-17DE102 ', b'8636 ', b'8621 ', b'7502 ', b'7840 ', b'477 ', b'7397 ', b'7043 ', b'f8619 ', b'7544 ', b'10084 ', b'7841 ', b'AI2600-19PL001 ', b'AD2700-18FR013 ', b'OIN-15-AR-DO-02 ', b'OIN-015-ARDP-08 ', b'OIN-15-ARDP-12 ', b'592 ', b'AL2500-17DE014 ', b'AI2632-19EU033 ', b'AL2500-17FR008 ', b'7583 ', b'8138 ', b'AI2600-17DE003 ', b'7186 ', b'AI2600-19PL001 ', b'AD2700-18NO003 ', b'1124 ', b'AI2600-18FR107 ', b'AI2632-18EU025 ', b'AI2600-19EU023 ', b'AI2600-20EU004 ', b'AI2632-19EU033 ', b'AI2600-16FR120 ', b'AI2632-19EU026 ', b'AI2600-16FR030 ', b'AI2632-19BU003 ', b'AI2600-19DE030 ', b'AI2600-19DE029 ', b'591 ', b'AI2600-18EU014 ', b'AI2600-18DE002 ', b'AI2600-19DE001 ', b'AL2500-17SP015 ', b'AL2500-18DE006 ', b'AI2600-19DE022 ', b'P32826-17FR003 ', b'AL2500-18DE004 ', b'AL2500-17SP003 ', b'P32826-17FR006 ', b'AI2600-19DE013 ', b'AL2500-17SP004 ', b'8726 ', b'P32826-17FR008 ', b'260020CA11 ', b'AL2500-16FR035 ', b'AL2500-17DE018 ', b'260020CA12 ', b'260019CA34 ', b'7702 ', b'260019CA38 ', b'AI2600-19SP001 ', b'260019CA30 ', b'8460 ', b'7355 ', b'260019CA39 ', b'7248 ', b'260018CA15 ', b'7836 ', b'AL2500-16FR029 ', b'7363 ', b'8461 ', b'AI2600-20EU004 ', b'7292 ', b'7418 ', b'AI2600-16FR126 ', b'P32826-19FR009 ', b'7562 ', b'P32826-19FR010 ', b'7233 ', b'7541 ', b'5355 ', b'7380 ', b'OIN-14-AR-65 ', b'AI2600-20EU004 '], dtype=object) - FIRMWARE_VERSION(N_PROF)object...
- long_name :
- Instrument firmware version
array([b'SBE602 11Dec15 ARM V2.1 ', b'SBE602 15Aug17 ARM V2.4 ', b'5608A15 ', b'030715 ', b'SBE602 11Dec15 ARM V2.1 ', b'121915 ', b'SBE602 20Sep13 ', b'5608A14 ', b'5900A04 ', b'5608A15 ', b'SBE602 11Dec15 ARM V2.1 ', b'102015 ', b'5608A15 ', b'5900A05 ', b'SBE602 11Dec15 ARM V2.1 ', b'SBE602 11Dec15 ARM V2.1 ', b'5900A04 ', b'5605A07 ', b'5900A04 ', b'5900A01 ', b'SBE602 11Dec15 ARM V2.1 ', b'5900A04 ', b'SBE602 11Dec15 ARM V2.1 ', b'5900A01 ', b'5900A04 ', b'060408 ', b'SBE602 15Aug17 ARM V2.4 ', b'2.11.0 ', b'SBE602 V1.3 ', b'5608A14 ', b'5900A05 ', b'SBE602 ARM_v2.0_xmsg_ve ', b'SBE603 01Dec16 ', b'6.0.1 ', b'SBE602 15Aug17 ARM V2.4 ', b'020717 ', b'6.0.1 ', b'5605B04 ', b'SBE602 11Dec15 ARM V2.1 ', b'5605B04 ', b'5900A01 ', b'SBE602 11Dec15 ARM V2.1 ', b'SBE603 01Dec16 ', b'SBE61v5.0.2 04May18 ', b'5900A05 ', b'3.01 ', b'5900A04 ', b'5900A00 ', b'5900A04 ', b'SBE602 ARM_v2.0_xmsg_ve ', b'SBE602 15Aug17 ARM V2.4 ', b'020117 ', b'3.01 ', b'081515 ', b'112015 ', b'020717 ', b'SBE602 15Aug17 ARM V2.4 ', b'6.0.1 ', b'SBE602 20Sep13 ', b'SBE602 11Dec15 ARM V2.1 ', b'5900A04 ', b'5900A04 ', b'5900A05 ', b'SBE602 15Aug17 ARM V2.4 ', b'5900A00 ', b'5900A04 ', b'5900A04 ', b'5900A04 ', b'5900A04 ', b'5900A04 ', b'37927 ', b'5900A00 ', b'5900A00 ', b'1.07 ', b'5900A01 ', b'5900A00 ', b'5900A00 ', b'SBE602 15Aug17 ARM V2.4 ', b'SBE602 11Dec15 ARM V2.1 ', b'3.02 ', b'SBE602 ARM_v2.0_xmsg_ve ', b'5608A11_26042018 ', b'121915 ', b'SBE602 11Dec15 ARM V2.1 ', b'5900A04 ', b'1.07 ', b'5900A04 ', b'2.13.1 ', b'2.14.3 ', b'5900A04 ', b'SBE602 15Aug17 ARM V2.4 ', b'SBE602 15Aug17 ARM V2.4 ', b'SBE603 01Dec16 ', b'2.6.1 ', b'1.07 ', b'5900A04 ', b'030716 ', b'5900A05 ', b'5900A04 ', b'020717 ', b'V2.0; SBE602 09Oct14 ', b'SBE602 11Dec15 ARM V2.1 ', b'SBE602 11Dec15 ARM V2.1 ', b'5605B04 ', b'37927 ', b'2.11.3.S ', b'SBE602 15Aug17 ARM V2.4 ', b'102015 ', b'n/a ', b'SBE602 11Dec15 ARM V2.1 ', b'SBE602 20Sep13 ', b'2.11.3 ', b'SBE602 15Aug17 ARM V2.4 ', b'SBE602 11Dec15 ARM V2.1 ', b'Apf9tSbe41.7.2.5-Flash-102015-BM', b'5900A05 ', b'5608A14 ', b'5605B05 ', b'5608A14 ', b'5608C08 ', b'n/a ', b'5605B04 ', b'5900A05 ', b'5605B04 ', b'112015 ', b'2.6.1 ', b'5900A04 ', b'SBE602 20Sep13 ', b'5900A05 ', b'5608A13 ', b'170425 ', b'5900A04 ', b'5900A04 ', b'5900A04 ', b'5900A05 ', b'5900A05 ', b'5900A01 ', b'5900A04 ', b'5900A00 ', b'5900A04 ', b'5900A04 ', b'5900A04 ', b'n/a ', b'5900A04 ', b'5900A04 ', b'5900A04 ', b'n/a ', b'5605B04 ', b'5900A04 ', b'5900A04 ', b'5605B04 ', b'n/a ', b'5900A04 ', b'5900A04 ', b'5605B04 ', b'081119 ', b'5900A04 ', b'n/a ', b'5605B04 ', b'5605B04 ', b'n/a ', b'n/a ', b'121915 ', b'n/a ', b'5900A04 ', b'n/a ', b'2.11.0 ', b'SBE602 ARM_v2.0_xmsg_ve ', b'n/a ', b'SBE602 20Sep13 ', b'n/a ', b'102015 ', b'5605B04 ', b'SBE602 11Dec15 ARM V2.1 ', b'2.11.0 ', b'5900A05 ', b'SBE602 ARM_v2.0_xmsg_ve ', b'SBE602 11Dec15 ARM V2.1 ', b'5900A01 ', b'5900A04 ', b'102815 ', b'5900A04 ', b'SBE602 20Sep13 ', b'SBE602 15Aug17 ARM V2.4 ', b'061810 ', b'SBE602 11Dec15 ARM V2.1 ', b'5611A01 ', b'5900A05 '], dtype=object) - WMO_INST_TYPE(N_PROF)object...
- long_name :
- Coded instrument type
- conventions :
- Argo reference table 8
array([b'854 ', b'854 ', b'838 ', b'846 ', b'854 ', b'846 ', b'853 ', b'838 ', b'844 ', b'838 ', b'854 ', b'846 ', b'838 ', b'844 ', b'854 ', b'854 ', b'844 ', b'844 ', b'844 ', b'844 ', b'854 ', b'844 ', b'854 ', b'844 ', b'844 ', b'846 ', b'854 ', b'846 ', b'854 ', b'838 ', b'844 ', b'854 ', b'874 ', b'873 ', b'854 ', b'846 ', b'873 ', b'844 ', b'854 ', b'844 ', b'844 ', b'854 ', b'862 ', b'874 ', b'844 ', b'836 ', b'844 ', b'844 ', b'844 ', b'854 ', b'854 ', b'846 ', b'836 ', b'863 ', b'846 ', b'846 ', b'854 ', b'873 ', b'854 ', b'854 ', b'844 ', b'844 ', b'844 ', b'854 ', b'844 ', b'844 ', b'844 ', b'844 ', b'844 ', b'844 ', b'846 ', b'844 ', b'844 ', b'836 ', b'844 ', b'844 ', b'844 ', b'854 ', b'854 ', b'836 ', b'854 ', b'838 ', b'846 ', b'854 ', b'844 ', b'836 ', b'844 ', b'846 ', b'846 ', b'844 ', b'854 ', b'854 ', b'874 ', b'846 ', b'836 ', b'844 ', b'846 ', b'844 ', b'844 ', b'846 ', b'853 ', b'854 ', b'854 ', b'844 ', b'846 ', b'846 ', b'854 ', b'846 ', b'865 ', b'854 ', b'854 ', b'846 ', b'854 ', b'854 ', b'846 ', b'844 ', b'838 ', b'844 ', b'838 ', b'838 ', b'865 ', b'844 ', b'844 ', b'844 ', b'846 ', b'846 ', b'844 ', b'854 ', b'844 ', b'838 ', b'863 ', b'844 ', b'844 ', b'844 ', b'844 ', b'844 ', b'844 ', b'844 ', b'844 ', b'844 ', b'844 ', b'844 ', b'865 ', b'844 ', b'844 ', b'844 ', b'844 ', b'844 ', b'844 ', b'841 ', b'844 ', b'844 ', b'841 ', b'844 ', b'844 ', b'846 ', b'841 ', b'844 ', b'844 ', b'844 ', b'844 ', b'844 ', b'846 ', b'844 ', b'844 ', b'844 ', b'846 ', b'854 ', b'844 ', b'854 ', b'844 ', b'846 ', b'844 ', b'854 ', b'846 ', b'844 ', b'854 ', b'854 ', b'844 ', b'841 ', b'846 ', b'841 ', b'854 ', b'854 ', b'846 ', b'854 ', b'844 ', b'844 '], dtype=object) - JULD(N_PROF)datetime64[ns]...
- long_name :
- Julian day (UTC) of the station relative to REFERENCE_DATE_TIME
- standard_name :
- time
- conventions :
- Relative julian days with decimal part (as parts of day)
- resolution :
- 0.0
- axis :
- T
array(['2020-11-11T23:51:52.000000000', '2020-11-11T23:42:19.000000000', '2020-11-11T23:36:00.000000000', '2020-11-11T23:17:33.000000000', '2020-11-11T23:06:22.999999744', '2020-11-11T23:02:02.000000000', '2020-11-11T22:56:34.000000256', '2020-11-11T22:34:00.000000256', '2020-11-11T22:09:00.000000000', '2020-11-11T22:08:00.000000000', '2020-11-11T21:46:15.999999744', '2020-11-11T21:27:47.000000000', '2020-11-11T21:14:00.000000000', '2020-11-11T21:02:20.000000000', '2020-11-11T20:58:31.999999744', '2020-11-11T20:56:08.000000256', '2020-11-11T20:52:30.000000000', '2020-11-11T20:51:00.000000000', '2020-11-11T20:49:30.000000256', '2020-11-11T20:46:20.000000000', '2020-11-11T20:18:53.000000000', '2020-11-11T19:47:30.000000000', '2020-11-11T19:46:33.000000256', '2020-11-11T19:43:30.000000000', '2020-11-11T19:31:30.000000000', '2020-11-11T19:30:58.000000000', '2020-11-11T19:23:58.000000000', '2020-11-11T18:33:58.000000000', '2020-11-11T18:28:50.000000000', '2020-11-11T18:12:00.000000256', '2020-11-11T18:01:20.000000256', '2020-11-11T17:59:08.000000000', '2020-11-11T17:58:34.000000000', '2020-11-11T17:34:50.000000000', '2020-11-11T17:33:02.000000000', '2020-11-11T17:26:21.000000256', '2020-11-11T17:23:58.000000000', '2020-11-11T17:00:00.000000000', '2020-11-11T16:59:11.000000000', '2020-11-11T16:56:00.000000000', '2020-11-11T16:53:30.000000000', '2020-11-11T16:20:05.000000256', '2020-11-11T16:02:24.000000256', '2020-11-11T15:31:21.000000000', '2020-11-11T15:06:20.000000000', '2020-11-11T15:00:00.000000000', '2020-11-11T14:57:30.000000256', '2020-11-11T14:41:30.000000256', '2020-11-11T14:38:30.000000000', '2020-11-11T14:28:52.999999744', '2020-11-11T14:02:32.000000000', '2020-11-11T13:59:04.000000000', '2020-11-11T13:54:00.000000000', '2020-11-11T13:33:33.000000256', '2020-11-11T13:17:24.000000000', '2020-11-11T13:13:22.999999744', '2020-11-11T13:04:35.000000000', '2020-11-11T13:03:47.000000000', '2020-11-11T12:48:21.000000256', '2020-11-11T12:30:45.000000000', '2020-11-11T12:08:30.000000000', '2020-11-11T12:05:00.000000000', '2020-11-11T12:01:20.000000256', '2020-11-11T11:56:52.000000000', '2020-11-11T11:55:30.000000000', '2020-11-11T11:54:29.999999744', '2020-11-11T11:54:20.000000000', '2020-11-11T11:52:20.000000000', '2020-11-11T11:50:20.000000000', '2020-11-11T11:46:30.000000000', '2020-11-11T11:43:45.000000000', '2020-11-11T11:43:30.000000000', '2020-11-11T11:41:30.000000256', '2020-11-11T11:39:59.999999744', '2020-11-11T11:38:30.000000000', '2020-11-11T11:35:30.000000000', '2020-11-11T11:34:30.000000000', '2020-11-11T11:32:57.000000000', '2020-11-11T11:18:12.999999744', '2020-11-11T11:17:00.000000000', '2020-11-11T11:05:31.999999744', '2020-11-11T11:05:30.000000000', '2020-11-11T11:04:21.000000000', '2020-11-11T11:01:33.000000000', '2020-11-11T10:50:00.000000256', '2020-11-11T10:39:00.000000000', '2020-11-11T10:33:00.000000000', '2020-11-11T10:16:50.000000256', '2020-11-11T10:07:35.000000000', '2020-11-11T10:07:30.000000000', '2020-11-11T10:04:57.000000000', '2020-11-11T10:03:32.000000000', '2020-11-11T09:51:13.000000000', '2020-11-11T09:40:00.000000000', '2020-11-11T09:34:00.000000000', '2020-11-11T09:32:30.000000000', '2020-11-11T09:25:59.000000256', '2020-11-11T09:01:20.000000256', '2020-11-11T08:55:59.999999744', '2020-11-11T08:50:45.000000000', '2020-11-11T08:48:40.000000000', '2020-11-11T08:47:36.000000000', '2020-11-11T08:20:08.000000000', '2020-11-11T08:20:00.000000000', '2020-11-11T08:14:35.000000256', '2020-11-11T08:07:09.000000000', '2020-11-11T08:03:03.000000000', '2020-11-11T08:00:52.000000000', '2020-11-11T07:54:59.999999744', '2020-11-11T07:46:26.000000000', '2020-11-11T07:36:08.000000000', '2020-11-11T07:26:38.000000000', '2020-11-11T07:18:23.000000000', '2020-11-11T07:18:09.000000000', '2020-11-11T07:02:20.000000256', '2020-11-11T06:59:00.000000000', '2020-11-11T06:59:00.000000000', '2020-11-11T06:58:00.000000000', '2020-11-11T06:56:00.000000000', '2020-11-11T06:55:20.000000000', '2020-11-11T06:52:00.000000000', '2020-11-11T06:49:03.000000000', '2020-11-11T06:46:00.000000000', '2020-11-11T06:40:59.999999744', '2020-11-11T06:39:59.000000000', '2020-11-11T06:38:40.000000000', '2020-11-11T06:32:59.999999744', '2020-11-11T06:30:53.000000000', '2020-11-11T06:18:20.000000000', '2020-11-11T06:14:00.000000000', '2020-11-11T06:09:41.000000000', '2020-11-11T06:03:30.000000000', '2020-11-11T06:03:30.000000000', '2020-11-11T06:00:30.000000000', '2020-11-11T06:00:20.000000000', '2020-11-11T05:55:30.000000000', '2020-11-11T05:55:20.000000000', '2020-11-11T05:50:30.000000000', '2020-11-11T05:49:30.000000256', '2020-11-11T05:49:00.000000000', '2020-11-11T05:48:20.000000256', '2020-11-11T05:47:20.000000000', '2020-11-11T05:41:00.000000000', '2020-11-11T05:39:59.999999744', '2020-11-11T05:39:00.000000000', '2020-11-11T05:38:20.000000000', '2020-11-11T05:37:00.000000000', '2020-11-11T05:35:00.000000256', '2020-11-11T05:33:20.000000000', '2020-11-11T05:32:30.000000000', '2020-11-11T05:31:00.000000000', '2020-11-11T05:27:00.000000256', '2020-11-11T05:26:30.000000000', '2020-11-11T05:26:20.000000000', '2020-11-11T05:24:00.000000000', '2020-11-11T05:16:48.000000000', '2020-11-11T05:13:30.000000000', '2020-11-11T05:13:00.000000000', '2020-11-11T05:06:00.000000000', '2020-11-11T05:05:00.000000000', '2020-11-11T04:57:00.000000000', '2020-11-11T04:54:59.999999744', '2020-11-11T04:53:37.999999744', '2020-11-11T04:49:00.000000000', '2020-11-11T04:48:20.000000000', '2020-11-11T04:48:00.000000000', '2020-11-11T04:47:49.000000000', '2020-11-11T04:47:27.000000000', '2020-11-11T04:46:00.000000000', '2020-11-11T04:28:04.000000000', '2020-11-11T04:25:00.000000000', '2020-11-11T04:17:51.999999744', '2020-11-11T04:17:00.000000000', '2020-11-11T03:59:00.000000000', '2020-11-11T03:03:01.000000000', '2020-11-11T03:02:20.000000000', '2020-11-11T02:55:25.000000256', '2020-11-11T02:51:42.000000000', '2020-11-11T02:49:30.000000256', '2020-11-11T02:46:30.000000000', '2020-11-11T02:23:14.000000000', '2020-11-11T02:04:30.000000256', '2020-11-11T02:01:25.000000000', '2020-11-11T01:59:01.000000000', '2020-11-11T01:40:56.000000000', '2020-11-11T01:23:27.000000000', '2020-11-11T01:01:59.999999744', '2020-11-11T00:08:20.000000000'], dtype='datetime64[ns]') - JULD_QC(N_PROF)object...
- long_name :
- Quality on date and time
- conventions :
- Argo reference table 2
array([b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1'], dtype=object) - JULD_LOCATION(N_PROF)datetime64[ns]...
- long_name :
- Julian day (UTC) of the location relative to REFERENCE_DATE_TIME
- conventions :
- Relative julian days with decimal part (as parts of day)
- resolution :
- 0.0
array(['2020-11-11T23:53:00.000000000', '2020-11-11T23:42:00.000000000', '2020-11-12T00:05:33.000000000', '2020-11-11T23:30:10.000000000', '2020-11-11T23:09:00.000000000', '2020-11-11T23:46:50.000000256', '2020-11-11T23:01:00.000000000', '2020-11-11T23:03:05.000000000', '2020-11-11T22:25:49.000000000', '2020-11-11T22:35:07.000000000', '2020-11-11T21:49:00.000000256', '2020-11-11T22:10:44.000000000', '2020-11-11T21:42:08.000000000', '2020-11-11T21:18:43.000000000', '2020-11-11T21:00:00.000000000', '2020-11-11T20:59:00.000000000', '2020-11-11T21:08:43.000000000', '2020-11-11T21:22:57.000000000', '2020-11-11T21:20:34.000000000', '2020-11-11T21:02:50.000000256', '2020-11-11T20:23:00.000000000', '2020-11-11T20:03:58.000000000', '2020-11-11T19:48:00.000000000', '2020-11-11T19:59:56.000000000', '2020-11-11T19:47:55.000000256', '2020-11-11T19:30:58.000000000', '2020-11-11T19:24:00.000000000', '2020-11-11T18:33:58.000000000', '2020-11-11T18:38:00.000000000', '2020-11-11T18:39:50.000000256', '2020-11-11T18:17:13.000000000', '2020-11-11T18:03:00.000000000', '2020-11-11T17:54:00.000000000', '2020-11-11T17:57:00.000000000', '2020-11-11T17:33:00.000000000', '2020-11-11T17:26:21.000000256', '2020-11-11T17:46:00.000000000', '2020-11-11T17:26:26.000000000', '2020-11-11T17:01:00.000000000', '2020-11-11T17:11:51.000000256', '2020-11-11T17:10:26.000000000', '2020-11-11T16:22:00.000000000', '2020-11-11T16:01:59.999999744', '2020-11-11T15:31:00.000000000', '2020-11-11T15:23:00.000000000', '2020-11-11T14:35:22.000000000', '2020-11-11T15:13:52.999999744', '2020-11-11T14:58:04.000000000', '2020-11-11T14:54:56.000000000', '2020-11-11T14:33:00.000000000', '2020-11-11T14:02:59.999999744', '2020-11-11T13:59:04.000000000', '2020-11-11T14:14:52.000000000', '2020-11-11T13:33:38.000000000', '2020-11-11T13:17:24.000000000', '2020-11-11T13:13:22.999999744', '2020-11-11T13:05:00.000000256', '2020-11-11T13:25:59.999999744', '2020-11-11T12:59:00.000000000', '2020-11-11T12:32:00.000000000', '2020-11-11T12:24:28.999999744', '2020-11-11T12:05:00.000000000', '2020-11-11T12:17:56.000000000', '2020-11-11T11:57:00.000000000', '2020-11-11T12:13:13.999999744', '2020-11-11T12:10:23.000000000', '2020-11-11T12:10:51.000000000', '2020-11-11T12:08:49.000000256', '2020-11-11T12:06:54.000000000', '2020-11-11T12:02:20.000000000', '2020-11-11T11:59:27.000000256', '2020-11-11T12:00:04.000000000', '2020-11-11T11:58:06.000000000', '2020-11-11T12:06:29.000000000', '2020-11-11T11:54:19.000000000', '2020-11-11T11:51:25.000000000', '2020-11-11T11:50:34.000000000', '2020-11-11T11:34:00.000000000', '2020-11-11T11:20:00.000000000', '2020-11-11T11:39:20.999999744', '2020-11-11T11:08:00.000000000', '2020-11-11T11:32:53.000000000', '2020-11-11T11:04:31.000000000', '2020-11-11T11:02:59.999999744', '2020-11-11T10:34:49.000000000', '2020-11-11T11:06:46.000000000', '2020-11-11T09:59:25.000000000', '2020-11-11T10:51:55.000000000', '2020-11-11T10:15:54.999999744', '2020-11-11T10:34:49.000000000', '2020-11-11T10:05:00.000000256', '2020-11-11T10:04:00.000000000', '2020-11-11T09:50:00.000000000', '2020-11-11T09:40:00.000000000', '2020-11-11T10:00:08.000000000', '2020-11-11T09:59:25.000000000', '2020-11-11T09:39:00.000000000', '2020-11-11T09:17:36.000000000', '2020-11-11T09:12:59.000000000', '2020-11-11T08:50:45.000000000', '2020-11-11T08:52:05.000000000', '2020-11-11T08:49:00.000000000', '2020-11-11T08:22:00.000000000', '2020-11-11T09:33:24.000000000', '2020-11-11T08:30:02.999999744', '2020-11-11T08:26:00.000000000', '2020-11-11T08:02:59.999999744', '2020-11-11T09:33:41.000000000', '2020-11-11T07:54:59.999999744', '2020-11-11T07:49:00.000000000', '2020-11-11T08:08:00.000000000', '2020-11-11T07:58:33.000000000', '2020-11-11T07:19:00.000000000', '2020-11-11T07:19:00.000000000', '2020-11-11T07:02:20.000000256', '2020-11-11T06:34:49.000000000', '2020-11-11T07:28:15.000000000', '2020-11-11T07:44:23.999999744', '2020-11-11T07:25:12.999999744', '2020-11-11T07:12:24.000000000', '2020-11-11T06:52:00.000000000', '2020-11-11T06:51:42.000000000', '2020-11-11T06:11:26.000000000', '2020-11-11T07:57:50.000000000', '2020-11-11T06:39:59.000000000', '2020-11-11T06:38:40.000000000', '2020-11-11T06:32:59.999999744', '2020-11-11T06:40:59.999999744', '2020-11-11T06:34:49.000000000', '2020-11-11T06:41:42.000000000', '2020-11-11T06:24:00.000000000', '2020-11-11T06:20:05.000000000', '2020-11-11T06:30:57.000000000', '2020-11-11T06:16:52.000000000', '2020-11-11T06:17:26.000000000', '2020-11-11T06:11:26.000000000', '2020-11-11T06:11:00.000000000', '2020-11-11T06:17:55.000000256', '2020-11-11T06:06:02.000000000', '2020-11-11T06:05:56.999999744', '2020-11-11T06:04:51.000000256', '2020-11-11T06:03:55.999999744', '2020-11-11T05:41:00.000000000', '2020-11-11T05:39:59.999999744', '2020-11-11T05:39:00.000000000', '2020-11-11T05:54:40.000000000', '2020-11-11T06:17:33.000000000', '2020-11-11T06:00:55.000000000', '2020-11-11T05:49:58.000000000', '2020-11-11T06:01:20.000000256', '2020-11-11T06:22:49.000000000', '2020-11-11T06:11:25.000000256', '2020-11-11T05:54:25.000000000', '2020-11-11T05:43:46.000000000', '2020-11-11T06:08:28.999999744', '2020-11-11T05:37:30.000000000', '2020-11-11T05:40:19.000000000', '2020-11-11T05:15:22.000000000', '2020-11-11T05:23:13.000000000', '2020-11-11T06:11:58.000000000', '2020-11-11T04:58:31.000000256', '2020-11-11T04:58:05.000000000', '2020-11-11T05:18:30.000000000', '2020-11-11T04:51:09.999999744', '2020-11-11T05:04:55.000000000', '2020-11-11T04:50:44.000000000', '2020-11-11T04:47:49.000000000', '2020-11-11T04:49:00.000000000', '2020-11-11T04:51:27.000000000', '2020-11-11T04:39:00.000000000', '2020-11-11T04:40:31.000000000', '2020-11-11T08:01:53.000000000', '2020-11-11T06:29:54.000000000', '2020-11-11T04:01:00.000000000', '2020-11-11T03:03:01.000000000', '2020-11-11T03:18:50.000000000', '2020-11-11T02:57:00.000000000', '2020-11-11T02:55:00.000000000', '2020-11-11T03:06:27.999999744', '2020-11-11T03:14:49.000000000', '2020-11-11T02:42:10.000000000', '2020-11-11T02:32:38.000000000', '2020-11-11T02:20:00.000000000', '2020-11-11T01:59:00.000000000', '2020-11-11T03:06:16.000000000', '2020-11-11T01:24:00.000000000', '2020-11-11T01:40:20.000000000', '2020-11-11T00:24:43.000000000'], dtype='datetime64[ns]') - LATITUDE(N_PROF)float64...
- long_name :
- Latitude of the station, best estimate
- standard_name :
- latitude
- units :
- degree_north
- valid_min :
- -90.0
- valid_max :
- 90.0
- axis :
- Y
array([-20.87581 , 26.04917 , 56.224208, 25.267 , 6.00987 , -32.282 , -47.74375 , 58.768142, 62.264573, 59.857033, 28.20956 , -15.098 , 58.86548 , 35.064548, 38.08667 , 47.01418 , 33.374083, 15.966 , 81.735 , -29.593263, -2.34639 , -8.965042, -0.43838 , -36.518753, -40.124863, 31.0612 , 24.81839 , -24.4121 , 3.48264 , 35.614093, 35.072108, 63.30427 , -26.04177 , 9.46176 , -16.31882 , -69.504 , 17.09767 , 5.482 , 40.22992 , -29.061 , 30.496247, 31.62384 , 21.74043 , -21.4816 , 35.076548, -22.961045, 66.03977 , -22.755448, 71.00046 , 52.45242 , -39.31072 , -40.4914 , -22.960258, -58.342 , 58.2522 , -71.067 , 38.06182 , -32.38966 , 29.3858 , -0.7086 , 35.490988, 73.022407, 35.082768, -13.07513 , 69.734125, 19.390237, 25.315313, 24.470218, 25.138683, -2.011093, 16.05094 , -17.924325, 14.340652, 42.321472, -15.060235, 20.470493, 19.084855, 2.57007 , 31.85924 , 71.315917, -24.06958 , 60.087683, -59.654 , 39.67386 , 58.836988, 36.643527, 57.068438, 74.25261 , 77.98792 , 58.836988, 25.88102 , 25.61855 , -27.49463 , -51.3824 , 34.20131 , 57.068438, 3.926 , 35.092623, 56.01898 , -69.621 , -58.8 , 5.53219 , 24.58431 , 31.366 , 42.40331 , 65.7023 , 16.01417 , 26.024 , 43.31319 , -32.59438 , 33.6996 , 67.91864 , 27.35783 , 22.40455 , 60.5225 , 54.753633, 48.013872, 56.945 , 49.082123, 43.805452, 45.349117, 71.408 , 55.147652, 15.458 , -42.9652 , -55.5627 , -60.9511 , 17.24889 , 54.753633, 78.15882 , -13.31 , 40.168738, 37.428652, 39.004813, 35.108467, 55.147652, 32.931428, 33.472492, -9.965365, 42.41645 , 0.191003, -0.278005, 36.901081, -67.532921, -61.969762, -17.761455, 54.296 , -9.911 , 14.011388, 35.193475, -10.019 , 25.024 , 33.454232, 36.892063, 33.395 , -51.648 , 37.453193, 41.421188, -34.67 , 20.317 , 42.32951 , 57.499676, -43.358 , 57.296295, 40.462665, 42.045486, 55.4008 , -33.16686 , 56.874912, 34.23417 , 59.204441, 43.518 , -28.509 , -16.35394 , 59.7019 , 35.127313, -24.45177 , 28.77227 , -0.62434 , 8.75631 , 65.392 , 43.373455, 19.79742 , 23.69158 , -48.749 , 34.49676 , 47.873 , 35.141292]) - LONGITUDE(N_PROF)float64...
- long_name :
- Longitude of the station, best estimate
- standard_name :
- longitude
- units :
- degree_east
- valid_min :
- -180.0
- valid_max :
- 180.0
- axis :
- X
array([-2.449676e+01, -8.620608e+01, -5.182488e+01, -5.208300e+01, -2.710274e+01, -2.538500e+01, -5.280813e+01, -5.266178e+01, 1.904156e+01, -5.354301e+01, -6.337559e+01, -3.436700e+01, -5.371946e+01, 1.575084e+01, -5.390500e+01, -2.717963e+01, 3.302149e+01, -3.945200e+01, 3.244447e+01, -1.154220e+01, -2.393146e+01, -3.299098e+01, -2.461274e+01, -6.857492e+00, -5.016751e+01, -5.723790e+01, -8.891927e+01, -2.863190e+01, -3.296480e+01, 2.011601e+01, 1.573373e+01, -3.092877e+01, -3.215065e+01, -5.287529e+01, -7.782340e+00, -3.691700e+01, -4.723997e+01, -1.181100e+01, -6.354800e+01, -6.500000e+00, -5.000990e+01, -6.546818e+01, -5.784617e+01, -2.362323e+01, 1.571878e+01, -2.770827e+01, -2.652797e+00, -1.155733e+01, 1.517156e+01, -2.612114e+01, -5.228520e+01, -1.444920e+01, -2.770735e+01, 1.090100e+01, -5.754800e+01, -2.984800e+01, -7.312869e+01, -1.154699e+01, -7.729474e+01, -1.598923e+01, -4.232118e+00, -6.685282e+01, 1.570761e+01, -3.525926e+01, -5.793550e+00, -9.482312e+01, -9.446385e+01, -9.152439e+01, -9.329708e+01, -3.986101e+01, -3.514955e+01, 6.840417e+00, -7.269610e+01, 6.614073e+00, -1.324221e+01, -4.113841e+01, -2.018198e+01, -2.642180e+01, -7.523737e+01, 1.224173e+01, -1.145608e+01, -3.971851e+01, -2.997100e+01, -2.670860e+01, 2.191978e+01, 1.641857e+00, 1.976775e+01, 7.163390e+00, 2.954717e+01, 2.191978e+01, -9.001734e+01, -8.632933e+01, -2.316034e+01, -5.919500e+00, 2.771104e+01, 1.976775e+01, -2.528000e+01, 1.570018e+01, -1.790219e+01, -2.494800e+01, -1.560000e+01, -2.323740e+01, -8.736726e+01, -4.371700e+01, 5.025870e+00, 3.118880e+00, -5.508360e+01, -7.670400e+01, -4.707825e+01, 1.071798e+01, -5.188640e+01, -1.253916e+01, -8.885741e+01, -9.526193e+01, -5.039770e+01, 1.888841e+01, -3.565567e+01, -4.203500e+01, -3.579873e+01, -1.735841e+01, -4.619118e+01, -1.872600e+01, 1.950122e+01, -2.490300e+01, -4.777160e+01, -1.963370e+01, 2.025700e+00, -2.134114e+01, 1.888841e+01, 7.977670e+00, -1.898200e+01, 1.065109e+01, 1.053477e+00, 7.050083e-01, 1.569738e+01, 1.950122e+01, 2.747595e+01, 3.503904e+01, -2.562341e+01, 3.329974e+01, -3.927854e+01, -4.171384e+01, -4.359353e+01, -1.399408e+01, -5.815235e+00, -3.703156e+01, -2.426400e+01, 5.977000e+00, -2.304328e+01, 1.648328e+01, -2.797700e+01, -2.096800e+01, 2.341561e+01, -2.827287e+01, -1.444100e+01, -4.738800e+01, 1.864481e+01, -6.317698e+01, 1.747900e+01, -3.095200e+01, -6.148556e+01, -5.167205e+01, -5.426300e+01, -5.330690e+01, 4.450870e+00, -6.114674e+01, -3.143790e+01, -3.872010e+00, -5.432794e+01, -7.430597e+01, -5.010341e+01, -5.537700e+01, -3.651400e+01, -5.767110e+00, -9.750300e+00, 1.569410e+01, -2.963033e+01, -7.528396e+01, -4.037098e+01, -5.851316e+01, -7.500000e-02, 7.446398e+00, -6.324159e+01, -8.240843e+01, 1.654500e+01, -6.374070e+01, -3.527300e+01, 1.568804e+01]) - POSITION_QC(N_PROF)object...
- long_name :
- Quality on position (latitude and longitude)
- conventions :
- Argo reference table 2
array([b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'2', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'8', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'8', b'1', b'8', b'1', b'1', b'1', b'1', b'1', b'8', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'8', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'8', b'8', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'8', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'8', b'8', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1', b'1'], dtype=object) - POSITIONING_SYSTEM(N_PROF)object...
- long_name :
- Positioning system
array([b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'ARGOS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'ARGOS ', b'IRIDIUM ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'ARGOS ', b'GPS ', b'ARGOS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'ARGOS ', b'GPS ', b'ARGOS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'ARGOS ', b'GPS ', b'GPS ', b'ARGOS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'ARGOS ', b'GPS ', b'GPS ', b'GPS ', b'ARGOS ', b'GPS ', b'GPS ', b'IRIDIUM ', b'GPS ', b'GPS ', b'GPS ', b'ARGOS ', b'GPS ', b'GPS ', b'ARGOS ', b'GPS ', b'GPS ', b'GPS ', b'ARGOS ', b'GPS ', b'ARGOS ', b'ARGOS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'ARGOS ', b'ARGOS ', b'GPS ', b'GPS ', b'ARGOS ', b'ARGOS ', b'GPS ', b'GPS ', b'ARGOS ', b'GPS ', b'GPS ', b'GPS ', b'ARGOS ', b'ARGOS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'ARGOS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'ARGOS ', b'ARGOS ', b'GPS ', b'ARGOS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'GPS ', b'ARGOS ', b'GPS ', b'ARGOS ', b'GPS '], dtype=object) - PROFILE_PRES_QC(N_PROF)object...
- long_name :
- Global quality flag of PRES profile
- conventions :
- Argo reference table 2a
array([b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'F', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'F', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A'], dtype=object) - PROFILE_TEMP_QC(N_PROF)object...
- long_name :
- Global quality flag of TEMP profile
- conventions :
- Argo reference table 2a
array([b'A', b'A', b'A', b'B', b'A', b'B', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'B', b'A', b'A', b'F', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'B', b'A', b'A', b'A', b'B', b'A', b'B', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'B', b'A', b'A', b'A', b'B', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'B', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'B', b'A', b'A', b'B', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'B', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'B', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'B', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'B', b'A', b'A', b'A', b'A', b'A', b'A', b'B', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'F', b'A', b'A', b'A', b'A', b'A', b'B', b'B', b'A', b'A', b'A', b'A', b'A', b'F', b'A', b'A'], dtype=object) - PROFILE_PSAL_QC(N_PROF)object...
- long_name :
- Global quality flag of PSAL profile
- conventions :
- Argo reference table 2a
array([b'F', b'A', b'A', b'F', b'F', b'B', b'A', b'A', b'A', b'A', b'F', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'F', b'F', b'A', b'A', b'A', b'A', b'A', b'B', b'A', b'A', b'F', b'A', b'A', b'A', b'B', b'A', b'A', b'F', b'A', b'A', b'A', b'B', b'A', b'A', b'A', b'B', b'A', b'B', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'F', b'A', b'A', b'A', b'B', b'F', b'F', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'F', b'A', b'B', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'B', b'A', b'A', b'B', b'F', b'F', b'A', b'A', b'A', b'A', b'A', b'A', b'B', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'B', b'A', b'A', b'A', b'A', b'F', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'B', b'A', b'A', b'A', b'A', b'B', b'A', b'A', b'A', b'F', b'A', b'A', b'A', b'A', b'A', b'F', b'A', b'A', b'F', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'B', b'A', b'A', b'F', b'A', b'A', b'A', b'F', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'A', b'F', b'A', b'A', b'A', b'F', b'F', b'B', b'B', b'A', b'A', b'A', b'A', b'A', b'F', b'A', b'A'], dtype=object) - VERTICAL_SAMPLING_SCHEME(N_PROF)object...
- long_name :
- Vertical sampling scheme
- conventions :
- Argo reference table 16
array([b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: averaged [10 sec sampling, 7 dbar average from 4000 dbar to 1400 dbar; 10 sec sampling, 5 dbar average from 1400 dbar to 400 dbar; 10 sec sampling, 1 dbar average from 400 dbar to 2.4 dbar] ', b'Primary sampling: mixed [deeper than nominal 985dbar: discrete; nominal 985dbar to surface: 2dbar-bin averaged] ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: mixed [deeper than nominal 985dbar: discrete; nominal 985dbar to surface: 2dbar-bin averaged] ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 1.0 Hz from a SBE41CP; bin detail from 0 dbar (number bins/bin width): 10/ 1;1005/ 2] ', b'Primary sampling: averaged [10 sec sampling, 7 dbar average from 4000 dbar to 1400 dbar; 10 sec sampling, 5 dbar average from 1400 dbar to 400 dbar; 10 sec sampling, 1 dbar average from 400 dbar to 2.0 dbar] ', b'Primary sampling: averaged [10 sec sampling, 1 dbar average from 105 dbar to 300 dbar; 10 sec sampling, 1 dbar average from 300 dbar to 10 dbar; 10 sec sampling, 1 dbar average from 10 dbar to 2.2 dbar] ', b'Primary sampling: averaged [10 sec sampling, 7 dbar average from 4000 dbar to 1400 dbar; 10 sec sampling, 5 dbar average from 1400 dbar to 400 dbar; 10 sec sampling, 1 dbar average from 400 dbar to 2.2 dbar] ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: discrete [] ', b'Primary sampling: averaged [10 sec sampling, 7 dbar average from 4000 dbar to 1400 dbar; 10 sec sampling, 5 dbar average from 1400 dbar to 400 dbar; 10 sec sampling, 1 dbar average from 400 dbar to 2.1 dbar] ', b'Primary sampling: averaged [10 sec sampling, 1 dbar average from 500 dbar to 700 dbar; 10 sec sampling, 1 dbar average from 700 dbar to 100 dbar; 10 sec sampling, 1 dbar average from 100 dbar to 2.5 dbar] ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: averaged [10 sec sampling, 25 dbar average from 2000 dbar to 700 dbar; 10 sec sampling, 10 dbar average from 700 dbar to 100 dbar; 10 sec sampling, 2 dbar average from 100 dbar to 2.3 dbar] ', b'Primary sampling: averaged [10 sec sampling, 25 dbar average from 2000 dbar to 200 dbar; 10 sec sampling, 10 dbar average from 200 dbar to 20 dbar; 10 sec sampling, 1 dbar average from 20 dbar to 5.5 dbar] ', b'Primary sampling: averaged [10 sec sampling, 5 dbar average from 2000 dbar to 500 dbar; 10 sec sampling, 2 dbar average from 500 dbar to 100 dbar; 10 sec sampling, 1 dbar average from 100 dbar to 2.2 dbar] ', b'Primary sampling: averaged [10 sec sampling, 5 dbar average from 2000 dbar to 500 dbar; 10 sec sampling, 2 dbar average from 500 dbar to 100 dbar; 10 sec sampling, 1 dbar average from 100 dbar to 2.6 dbar] ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: averaged [10 sec sampling, 5 dbar average from 2000 dbar to 500 dbar; 10 sec sampling, 2 dbar average from 500 dbar to 100 dbar; 10 sec sampling, 1 dbar average from 100 dbar to 2.2 dbar] ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: averaged [10 sec sampling, 5 dbar average from 2000 dbar to 500 dbar; 10 sec sampling, 2 dbar average from 500 dbar to 100 dbar; 10 sec sampling, 1 dbar average from 100 dbar to 2.2 dbar] ', b'Primary sampling: averaged [10 sec sampling, 5 dbar average from 2000 dbar to 500 dbar; 10 sec sampling, 2 dbar average from 500 dbar to 100 dbar; 10 sec sampling, 1 dbar average from 100 dbar to 2.2 dbar] ', b'Primary sampling: discrete ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: discrete ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: averaged [10 s sampling, 1 dbar avg from 4000 dbar to 3049.4 dbar;10 s sampling, 25 dbar avg from 3049.4 dbar to 700 dbar;10 s sampling, 10 dbar avg from 700 dbar to 100 dbar;10 s sampling, 2 dbar avg from 100 dbar to 5.5 dbar] ', b'Primary sampling: averaged [10 sec sampling, 1 dbar average from 500 dbar to 700 dbar; 10 sec sampling, 1 dbar average from 700 dbar to 100 dbar; 10 sec sampling, 1 dbar average from 100 dbar to 2.3 dbar] ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: averaged [1Hz sampling by SBE-41CP averaged in 2-dbar bins] ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: mixed [deeper than nominal 985dbar: discrete; nominal 985dbar to surface: 2dbar-bin averaged] ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: averaged [10 sec sampling, 25 dbar average from 2000 dbar to 200 dbar; 10 sec sampling, 10 dbar average from 200 dbar to 10 dbar; 10 sec sampling, 1 dbar average from 10 dbar to 5.5 dbar] ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: averaged [10 sec sampling, 25 dbar average from 2000 dbar to 200 dbar; 10 sec sampling, 10 dbar average from 200 dbar to 10 dbar; 10 sec sampling, 1 dbar average from 10 dbar to 5.5 dbar] ', b'Primary sampling: averaged [10 sec sampling, 4 dbar average from 2000 dbar to 500 dbar; 10 sec sampling, 2 dbar average from 500 dbar to 100 dbar; 10 sec sampling, 1 dbar average from 100 dbar to 2.0 dbar] ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: mixed [PRES <= 510 dbar: nominal 2 dbar binned data sampled at 0.5 Hz from a SBE61, PRES > 510 dbar discrete sampling from the same SBE61] ', b'Primary sampling: averaged [1Hz sampling by SBE-41CP averaged in 2-dbar bins] ', b'Primary sampling: averaged [10 sec sampling, 1 dbar average from 500 dbar to 700 dbar; 10 sec sampling, 1 dbar average from 700 dbar to 100 dbar; 10 sec sampling, 1 dbar average from 100 dbar to 2.1 dbar] ', b'Primary sampling: averaged [60sec sampling, 1dbar average in [surface-1dbar]] ', b'Primary sampling: averaged [10 sec sampling, 5 dbar average from 2000 dbar to 500 dbar; 10 sec sampling, 2 dbar average from 500 dbar to 100 dbar; 10 sec sampling, 1 dbar average from 100 dbar to 2.0 dbar] ', b'Primary sampling: averaged [10 sec sampling, 5 dbar average from 2000 dbar to 500 dbar; 10 sec sampling, 2 dbar average from 500 dbar to 100 dbar; 10 sec sampling, 1 dbar average from 100 dbar to 2.4 dbar] ', b'Primary sampling: averaged [10 sec sampling, 5 dbar average from 2000 dbar to 500 dbar; 10 sec sampling, 2 dbar average from 500 dbar to 100 dbar; 10 sec sampling, 1 dbar average from 100 dbar to 2.1 dbar] ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: discrete ', b'Primary sampling: averaged [2s sampling,10dbar average in [1000-250]dbar;2s samp.,2dbar avg in [250-10]dbar;2s samp.,1dbar avg in [10-2.1]dbar] ', b'Primary sampling: mixed [deeper than nominal 985dbar: discrete; nominal 985dbar to surface: 2dbar-bin averaged] ', b'Primary sampling: discrete ', b'Primary sampling: mixed [deeper than nominal 985dbar: discrete; nominal 985dbar to surface: 2dbar-bin averaged] ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: averaged [10 sec sampling, 5 dbar average from 800 dbar to 1400 dbar; 10 sec sampling, 2 dbar average from 1400 dbar to 400 dbar; 10 sec sampling, 1 dbar average from 400 dbar to 2.7 dbar] ', b'Primary sampling: averaged [10 sec sampling, 25 dbar average from 1000 dbar to 1000 dbar; 10 sec sampling, 5 dbar average from 1000 dbar to 50 dbar; 10 sec sampling, 1 dbar average from 50 dbar to 2.5 dbar] ', b'Primary sampling: averaged [10 sec sampling, 1 dbar average from 500 dbar to 700 dbar; 10 sec sampling, 1 dbar average from 700 dbar to 100 dbar; 10 sec sampling, 1 dbar average from 100 dbar to 2.0 dbar] ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: averaged [10 sec sampling, 4 dbar average from 2000 dbar to 1400 dbar; 10 sec sampling, 2 dbar average from 1400 dbar to 250 dbar; 10 sec sampling, 1 dbar average from 250 dbar to 2.1 dbar] ', b'Primary sampling: averaged [10 sec sampling, 5 dbar average from 2000 dbar to 1400 dbar; 10 sec sampling, 2 dbar average from 1400 dbar to 400 dbar; 10 sec sampling, 1 dbar average from 400 dbar to 2.4 dbar] ', b'Primary sampling: averaged [10 sec sampling, 5 dbar average from 2000 dbar to 1400 dbar; 10 sec sampling, 2 dbar average from 1400 dbar to 400 dbar; 10 sec sampling, 1 dbar average from 400 dbar to 2.2 dbar] ', b'Primary sampling: averaged [10 sec sampling, 5 dbar average from 2000 dbar to 1400 dbar; 10 sec sampling, 2 dbar average from 1400 dbar to 400 dbar; 10 sec sampling, 1 dbar average from 400 dbar to 2.5 dbar] ', b'Primary sampling: averaged [10 sec sampling, 5 dbar average from 2000 dbar to 1400 dbar; 10 sec sampling, 2 dbar average from 1400 dbar to 400 dbar; 10 sec sampling, 1 dbar average from 400 dbar to 2.0 dbar] ', b'Primary sampling: averaged [10 sec sampling, 5 dbar average from 2000 dbar to 1400 dbar; 10 sec sampling, 2 dbar average from 1400 dbar to 400 dbar; 10 sec sampling, 1 dbar average from 400 dbar to 2.4 dbar] ', b'Primary sampling: mixed [averaged from 4.38 dbar to 743.31 dbar {[1 sample/second, 2 dbar average from 750 dbar to 4 dbar] for CTD; [1 second interval from 750 dbar to 4 dbar] for TRANSISTOR_PH}; discrete otherwise] ', b'Primary sampling: averaged [10 sec sampling, 5 dbar average from 2000 dbar to 500 dbar; 10 sec sampling, 2 dbar average from 500 dbar to 100 dbar; 10 sec sampling, 1 dbar average from 100 dbar to 2.1 dbar] ', b'Primary sampling: averaged [10 sec sampling, 25 dbar average from 2000 dbar to 200 dbar; 10 sec sampling, 10 dbar average from 200 dbar to 10 dbar; 10 sec sampling, 1 dbar average from 10 dbar to 2.5 dbar] ', b'Primary sampling: averaged [2s sampling,2dbar average in [1000-250]dbar;2s samp.,1dbar avg in [250-10]dbar;2s samp.,1dbar avg in [10-2.4]dbar] ', b'Primary sampling: averaged [10 sec sampling, 4 dbar average from 2000 dbar to 1400 dbar; 10 sec sampling, 2 dbar average from 1400 dbar to 250 dbar; 10 sec sampling, 1 dbar average from 250 dbar to 2.1 dbar] ', b'Primary sampling: averaged [10 sec sampling, 4 dbar average from 2000 dbar to 1400 dbar; 10 sec sampling, 2 dbar average from 1400 dbar to 250 dbar; 10 sec sampling, 1 dbar average from 250 dbar to 2.7 dbar] ', b'Primary sampling: averaged [10 sec sampling, 4 dbar average from 2000 dbar to 1400 dbar; 10 sec sampling, 2 dbar average from 1400 dbar to 250 dbar; 10 sec sampling, 1 dbar average from 250 dbar to 2.2 dbar] ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: averaged [2s sampling,5dbar average in [2000-1100]dbar;2s samp.,2dbar avg in [1100-300]dbar;2s samp.,2dbar avg in [300-10]dbar;2s samp.,1dbar avg in [10-2.3]dbar] ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: averaged [10 sec sampling, 7 dbar average from 4000 dbar to 1400 dbar; 10 sec sampling, 5 dbar average from 1400 dbar to 400 dbar; 10 sec sampling, 1 dbar average from 400 dbar to 2.3 dbar] ', b'Primary sampling: mixed [deeper than nominal 985dbar: discrete; nominal 985dbar to surface: 2dbar-bin averaged] ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: averaged [10 sec sampling, 1 dbar average from surface to 40 dbar; 10 sec sampling, 1 dbar average from 40 dbar to 80 dbar; 10 sec sampling, 1 dbar average from 80 dbar to 60 dbar] ', b'Primary sampling: averaged [2s sampling,2dbar average in [1000-250]dbar;2s samp.,1dbar avg in [250-10]dbar;2s samp.,1dbar avg in [10-2.1]dbar] ', b'Primary sampling: averaged [10 sec sampling, 1 dbar average from surface to 100 dbar; 10 sec sampling, 1 dbar average from 100 dbar to 200 dbar; 10 sec sampling, 1 dbar average from 200 dbar to 200 dbar] ', b'Primary sampling: averaged [1 sample/sec,2dbar avg from 2100 to 4dbar] ', b'Primary sampling: averaged [1 sample/sec,2dbar avg from 470 to 4dbar] ', b'Primary sampling: averaged [10 sec sampling, 1 dbar average from 200 dbar to 80 dbar; 10 sec sampling, 1 dbar average from 80 dbar to 40 dbar; 10 sec sampling, 1 dbar average from 40 dbar to 2.5 dbar] ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: averaged [1Hz sampling by SBE-41CP averaged in 2-dbar bins] ', b'Primary sampling: discrete ', b'Primary sampling: averaged [2s sampling,2dbar average in [1000-250]dbar;2s samp.,1dbar avg in [250-10]dbar;2s samp.,1dbar avg in [10-2.2]dbar] ', b'Primary sampling: averaged [10 sec sampling, 1 dbar average from 200 dbar to 200 dbar; 10 sec sampling, 1 dbar average from 200 dbar to 100 dbar; 10 sec sampling, 1 dbar average from 100 dbar to 2.5 dbar] ', b'Primary sampling: mixed [deeper than nominal 985dbar: discrete; nominal 985dbar to surface: 2dbar-bin averaged] ', b'Primary sampling: averaged [10 sec sampling, 1 dbar average from 500 dbar to 700 dbar; 10 sec sampling, 1 dbar average from 700 dbar to 100 dbar; 10 sec sampling, 1 dbar average from 100 dbar to 2.6 dbar] ', b'Primary sampling: averaged [10 sec sampling, 5 dbar average from 2000 dbar to 500 dbar; 10 sec sampling, 2 dbar average from 500 dbar to 100 dbar; 10 sec sampling, 1 dbar average from 100 dbar to 2.7 dbar] ', b'Primary sampling: mixed [deeper than nominal 985dbar: discrete; nominal 985dbar to surface: 2dbar-bin averaged] ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 1.0 Hz from a SBE41CP; bin detail from 0 dbar (number bins/bin width): 10/ 1;1005/ 2] ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: averaged [10 sec sampling, 20 dbar average from 2000 dbar to 200 dbar; 10 sec sampling, 10 dbar average from 200 dbar to 10 dbar; 10 sec sampling, 1 dbar average from 10 dbar to 5.5 dbar] ', b'Primary sampling: mixed [averaged from 2.55 dbar to 35.42 dbar {1 sample/sec,2dbar avg from 1000 to 4dbar}; discrete otherwise] ', b'Primary sampling: averaged [1 sample/sec,2dbar avg from 2100 to 4dbar] ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: discrete [] ', b'Primary sampling: discrete ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: averaged [1 sample/sec,2dbar avg from 2100 to 4dbar] ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: discrete ', b'Primary sampling: averaged [10 sec sampling, 1 dbar average from surface to 40 dbar; 10 sec sampling, 1 dbar average from 40 dbar to 80 dbar; 10 sec sampling, 1 dbar average from 80 dbar to 150 dbar] ', b'Primary sampling: averaged [10 sec sampling, 7 dbar average from 4000 dbar to 1400 dbar; 10 sec sampling, 5 dbar average from 1400 dbar to 400 dbar; 10 sec sampling, 1 dbar average from 400 dbar to 2.0 dbar] ', b'Primary sampling: averaged [10 sec sampling, 25 dbar average from 2000 dbar to 500 dbar; 10 sec sampling, 10 dbar average from 500 dbar to 10 dbar; 10 sec sampling, 1 dbar average from 10 dbar to 2.5 dbar] ', b'Primary sampling: averaged [10 sec sampling, 7 dbar average from 4000 dbar to 1400 dbar; 10 sec sampling, 5 dbar average from 1400 dbar to 400 dbar; 10 sec sampling, 1 dbar average from 400 dbar to 2.9 dbar] ', b'Primary sampling: averaged [10 sec sampling, 20 dbar average from 4000 dbar to 500 dbar; 10 sec sampling, 10 dbar average from 500 dbar to 10 dbar; 10 sec sampling, 1 dbar average from 10 dbar to 2.6 dbar] ', b'Primary sampling: discrete ', b'Primary sampling: averaged [10 sec sampling, 20 dbar average from 2000 dbar to 200 dbar; 10 sec sampling, 10 dbar average from 200 dbar to 10 dbar; 10 sec sampling, 1 dbar average from 10 dbar to 5.5 dbar] ', b'Primary sampling: averaged [8 sec sampling, 1 dbar average from surface to 10 dbar; 8 sec sampling, 1 dbar average from 10 dbar to 100 dbar; 8 sec sampling, 1 dbar average from 100 dbar to 80 dbar] ', b'Primary sampling: averaged [10 sec sampling, 25 dbar average from 2000 dbar to 200 dbar; 10 sec sampling, 10 dbar average from 200 dbar to 10 dbar; 10 sec sampling, 1 dbar average from 10 dbar to 2.5 dbar] ', b'Primary sampling: discrete ', b'Primary sampling: discrete ', b'Primary sampling: averaged [10 sec sampling, 25 dbar average from 2000 dbar to 200 dbar; 10 sec sampling, 10 dbar average from 200 dbar to 10 dbar; 10 sec sampling, 1 dbar average from 10 dbar to 5.5 dbar] ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: averaged [10 sec sampling, 1 dbar average from 200 dbar to 80 dbar; 10 sec sampling, 1 dbar average from 80 dbar to 40 dbar; 10 sec sampling, 1 dbar average from 40 dbar to 2.1 dbar] ', b'Primary sampling: averaged [10 sec sampling, 5 dbar average from 3500 dbar to 1000 dbar; 10 sec sampling, 2 dbar average from 1000 dbar to 10 dbar; 10 sec sampling, 1 dbar average from 10 dbar to 5.0 dbar] ', b'Primary sampling: averaged ', b'Primary sampling: averaged [10 sec sampling, 5 dbar average from 800 dbar to 1400 dbar; 10 sec sampling, 2 dbar average from 1400 dbar to 400 dbar; 10 sec sampling, 1 dbar average from 400 dbar to 2.8 dbar] ', b'Primary sampling: averaged [10 sec sampling, 25 dbar average from 700 dbar to 700 dbar; 10 sec sampling, 10 dbar average from 700 dbar to 100 dbar; 10 sec sampling, 2 dbar average from 100 dbar to 2.0 dbar] ', b'Primary sampling: averaged [10 sec sampling, 5 dbar average from 2000 dbar to 1400 dbar; 10 sec sampling, 2 dbar average from 1400 dbar to 400 dbar; 10 sec sampling, 1 dbar average from 400 dbar to 2.5 dbar] ', b'Primary sampling: averaged [10 sec sampling, 1 dbar average from 500 dbar to 700 dbar; 10 sec sampling, 1 dbar average from 700 dbar to 100 dbar; 10 sec sampling, 1 dbar average from 100 dbar to 2.1 dbar] ', b'Primary sampling: averaged [8 sec sampling, 1 dbar average from 200 dbar to 100 dbar; 8 sec sampling, 1 dbar average from 100 dbar to 10 dbar; 8 sec sampling, 1 dbar average from 10 dbar to 2.7 dbar] ', b'Primary sampling: averaged [10 sec sampling, 25 dbar average from 700 dbar to 700 dbar; 10 sec sampling, 10 dbar average from 700 dbar to 100 dbar; 10 sec sampling, 2 dbar average from 100 dbar to 2.8 dbar] ', b'Primary sampling: averaged [10 sec sampling, 25 dbar average from 2000 dbar to 700 dbar; 10 sec sampling, 10 dbar average from 700 dbar to 100 dbar; 10 sec sampling, 2 dbar average from 100 dbar to 2.9 dbar] ', b'Primary sampling: averaged [10 sec sampling, 5 dbar average from 2000 dbar to 500 dbar; 10 sec sampling, 2 dbar average from 500 dbar to 100 dbar; 10 sec sampling, 1 dbar average from 100 dbar to 2.1 dbar] ', b'Primary sampling: averaged [10 sec sampling, 25 dbar average from 2000 dbar to 200 dbar; 10 sec sampling, 10 dbar average from 200 dbar to 10 dbar; 10 sec sampling, 1 dbar average from 10 dbar to 5.0 dbar] ', b'Primary sampling: averaged [10 sec sampling, 25 dbar average from 2000 dbar to 200 dbar; 10 sec sampling, 10 dbar average from 200 dbar to 10 dbar; 10 sec sampling, 1 dbar average from 10 dbar to 5.7 dbar] ', b'Primary sampling: averaged [10 sec sampling, 25 dbar average from 2000 dbar to 200 dbar; 10 sec sampling, 10 dbar average from 200 dbar to 10 dbar; 10 sec sampling, 1 dbar average from 10 dbar to 5.2 dbar] ', b'Primary sampling: discrete ', b'Primary sampling: averaged [10 sec sampling, 25 dbar average from 2000 dbar to 700 dbar; 10 sec sampling, 10 dbar average from 700 dbar to 100 dbar; 10 sec sampling, 2 dbar average from 100 dbar to 2.5 dbar] ', b'Primary sampling: averaged [10 sec sampling, 25 dbar average from 2000 dbar to 200 dbar; 10 sec sampling, 10 dbar average from 200 dbar to 10 dbar; 10 sec sampling, 1 dbar average from 10 dbar to 5.5 dbar] ', b'Primary sampling: averaged [10 sec sampling, 25 dbar average from 2000 dbar to 200 dbar; 10 sec sampling, 10 dbar average from 200 dbar to 10 dbar; 10 sec sampling, 1 dbar average from 10 dbar to 5.6 dbar] ', b'Primary sampling: averaged [10 sec sampling, 25 dbar average from 2000 dbar to 200 dbar; 10 sec sampling, 10 dbar average from 200 dbar to 10 dbar; 10 sec sampling, 1 dbar average from 10 dbar to 5.5 dbar] ', b'Primary sampling: averaged [10 sec sampling, 20 dbar average from 2000 dbar to 200 dbar; 10 sec sampling, 10 dbar average from 200 dbar to 10 dbar; 10 sec sampling, 1 dbar average from 10 dbar to 5.5 dbar] ', b'Primary sampling: averaged [10 sec sampling, 25 dbar average from 2000 dbar to 200 dbar; 10 sec sampling, 10 dbar average from 200 dbar to 10 dbar; 10 sec sampling, 1 dbar average from 10 dbar to 5.4 dbar] ', b'Primary sampling: averaged [10 sec sampling, 5 dbar average from 2000 dbar to 1400 dbar; 10 sec sampling, 2 dbar average from 1400 dbar to 400 dbar; 10 sec sampling, 1 dbar average from 400 dbar to 2.1 dbar] ', b'Primary sampling: averaged [10 sec sampling, 20 dbar average from 2000 dbar to 200 dbar; 10 sec sampling, 10 dbar average from 200 dbar to 10 dbar; 10 sec sampling, 1 dbar average from 10 dbar to 5.5 dbar] ', b'Primary sampling: averaged [10 sec sampling, 25 dbar average from 2000 dbar to 200 dbar; 10 sec sampling, 10 dbar average from 200 dbar to 10 dbar; 10 sec sampling, 1 dbar average from 10 dbar to 5.5 dbar] ', b'Primary sampling: averaged [10 sec sampling, 5 dbar average from 2000 dbar to 1400 dbar; 10 sec sampling, 2 dbar average from 1400 dbar to 400 dbar; 10 sec sampling, 1 dbar average from 400 dbar to 2.3 dbar] ', b'Primary sampling: averaged [10 sec sampling, 25 dbar average from 2000 dbar to 200 dbar; 10 sec sampling, 10 dbar average from 200 dbar to 10 dbar; 10 sec sampling, 1 dbar average from 10 dbar to 5.5 dbar] ', b'Primary sampling: averaged [10 sec sampling, 25 dbar average from 2000 dbar to 200 dbar; 10 sec sampling, 10 dbar average from 200 dbar to 10 dbar; 10 sec sampling, 1 dbar average from 10 dbar to 5.5 dbar] ', b'Primary sampling: mixed [deeper than nominal 985dbar: discrete; nominal 985dbar to surface: 2dbar-bin averaged] ', b'Primary sampling: averaged [10 sec sampling, 5 dbar average from 2000 dbar to 1400 dbar; 10 sec sampling, 2 dbar average from 1400 dbar to 400 dbar; 10 sec sampling, 1 dbar average from 400 dbar to 2.8 dbar] ', b'Primary sampling: discrete ', b'Primary sampling: averaged [10 sec sampling, 25 dbar average from 2000 dbar to 200 dbar; 10 sec sampling, 10 dbar average from 200 dbar to 10 dbar; 10 sec sampling, 1 dbar average from 10 dbar to 5.5 dbar] ', b'Primary sampling: averaged [10 sec sampling, 20 dbar average from 2000 dbar to 200 dbar; 10 sec sampling, 10 dbar average from 200 dbar to 10 dbar; 10 sec sampling, 1 dbar average from 10 dbar to 5.5 dbar] ', b'Primary sampling: discrete ', b'Primary sampling: discrete ', b'Primary sampling: mixed [deeper than nominal 985dbar: discrete; nominal 985dbar to surface: 2dbar-bin averaged] ', b'Primary sampling: discrete ', b'Primary sampling: averaged [10 sec sampling, 5 dbar average from 2000 dbar to 1400 dbar; 10 sec sampling, 2 dbar average from 1400 dbar to 400 dbar; 10 sec sampling, 1 dbar average from 400 dbar to 2.0 dbar] ', b'Primary sampling: discrete ', b'Primary sampling: discrete ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: discrete ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: discrete ', b'Primary sampling: discrete [] ', b'Primary sampling: averaged [10 sec sampling, 25 dbar average from 2000 dbar to 200 dbar; 10 sec sampling, 10 dbar average from 200 dbar to 10 dbar; 10 sec sampling, 1 dbar average from 10 dbar to 5.5 dbar] ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: discrete ', b'Primary sampling: averaged [10 sec sampling, 1 dbar average from 500 dbar to 700 dbar; 10 sec sampling, 1 dbar average from 700 dbar to 100 dbar; 10 sec sampling, 1 dbar average from 100 dbar to 2.2 dbar] ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: averaged [10 sec sampling, 4 dbar average from 2000 dbar to 500 dbar; 10 sec sampling, 2 dbar average from 500 dbar to 250 dbar; 10 sec sampling, 1 dbar average from 250 dbar to 2.5 dbar] ', b'Primary sampling: averaged [10 sec sampling, 5 dbar average from 2000 dbar to 1400 dbar; 10 sec sampling, 2 dbar average from 1400 dbar to 1000 dbar; 10 sec sampling, 1 dbar average from 1000 dbar to 2.2 dbar] ', b'Primary sampling: averaged [2dbar-bin averaged] ', b'Primary sampling: averaged [10 sec sampling, 5 dbar average from 2000 dbar to 1400 dbar; 10 sec sampling, 2 dbar average from 1400 dbar to 400 dbar; 10 sec sampling, 1 dbar average from 400 dbar to 2.3 dbar] ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: discrete [] ', b'Primary sampling: averaged [nominal 2 dbar binned data sampled at 0.5 Hz from a SBE41CP] ', b'Primary sampling: averaged [10 sec sampling, 25 dbar average from 2000 dbar to 500 dbar; 10 sec sampling, 10 dbar average from 500 dbar to 10 dbar; 10 sec sampling, 1 dbar average from 10 dbar to 2.5 dbar] ', b'Primary sampling: averaged [10 sec sampling, 1 dbar average from 500 dbar to 700 dbar; 10 sec sampling, 1 dbar average from 700 dbar to 100 dbar; 10 sec sampling, 1 dbar average from 100 dbar to 2.4 dbar] '], dtype=object) - CONFIG_MISSION_NUMBER(N_PROF)float64...
- long_name :
- Unique number denoting the missions performed by the float
- conventions :
- 1...N, 1 : first complete mission
array([ 4., 3., 2., 180., 5., 113., 31., 2., 13., 2., 6., 2., 2., 2., 9., 4., 2., 2., 5., 2., 7., 2., 5., 2., 2., -1., 3., -1., 5., 3., 2., 4., 14., 6., 4., 103., 4., 2., 4., 2., 2., 5., 7., 9., 2., 13., 2., 4., 2., 5., 4., -1., 14., 69., -1., 103., 3., 8., 5., 6., 2., 4., 2., 3., 4., 4., 4., 4., 4., 5., 9., 4., 5., 4., 3., 3., 3., 4., 6., 4., 6., 4., 62., 4., 8., 9., 6., 4., 9., 8., 2., 3., 9., 23., 9., 6., 114., 2., 2., 103., 13., 5., 4., 2., 3., 7., 3., 2., 1., 5., 5., 2., 2., 6., -1., 5., 2., 2., 2., 2., 1., 2., 2., 2., -1., 52., 3., 2., 5., 2., 33., 2., 7., 2., 2., 2., 3., 2., 4., 2., 2., 2., 1., 2., 2., 2., 2., 2., 2., 4., 2., 3., 3., 2., 3., 31., 4., 1., 2., 2., 1., 1., 136., 1., 7., 1., -1., 5., 1., 5., 1., 2., 2., 4., -1., 2., 6., 4., 4., 4., 31., 2., 5., 3., 1., 9., 2., 2.]) - PRES(N_PROF, N_LEVELS)float32...
- long_name :
- Sea water pressure, equals 0 at sea-level
- standard_name :
- sea_water_pressure
- units :
- decibar
- valid_min :
- 0.0
- valid_max :
- 12000.0
- C_format :
- %7.1f
- FORTRAN_format :
- F7.1
- resolution :
- 1.0
- axis :
- Z
[250228 values with dtype=float32]
- PRES_QC(N_PROF, N_LEVELS)object...
- long_name :
- quality flag
- conventions :
- Argo reference table 2
[250228 values with dtype=object]
- PRES_ADJUSTED(N_PROF, N_LEVELS)float32...
- long_name :
- Sea water pressure, equals 0 at sea-level
- standard_name :
- sea_water_pressure
- units :
- decibar
- valid_min :
- 0.0
- valid_max :
- 12000.0
- C_format :
- %7.1f
- FORTRAN_format :
- F7.1
- resolution :
- 1.0
- axis :
- Z
[250228 values with dtype=float32]
- PRES_ADJUSTED_QC(N_PROF, N_LEVELS)object...
- long_name :
- quality flag
- conventions :
- Argo reference table 2
[250228 values with dtype=object]
- PRES_ADJUSTED_ERROR(N_PROF, N_LEVELS)float32...
- long_name :
- Contains the error on the adjusted values as determined by the delayed mode QC process
- units :
- decibar
- C_format :
- %7.1f
- FORTRAN_format :
- F7.1
- resolution :
- 1.0
[250228 values with dtype=float32]
- TEMP(N_PROF, N_LEVELS)float32...
- long_name :
- Sea temperature in-situ ITS-90 scale
- standard_name :
- sea_water_temperature
- units :
- degree_Celsius
- valid_min :
- -2.5
- valid_max :
- 40.0
- C_format :
- %9.3f
- FORTRAN_format :
- F9.3
- resolution :
- 0.001
[250228 values with dtype=float32]
- TEMP_QC(N_PROF, N_LEVELS)object...
- long_name :
- quality flag
- conventions :
- Argo reference table 2
[250228 values with dtype=object]
- TEMP_ADJUSTED(N_PROF, N_LEVELS)float32...
- long_name :
- Sea temperature in-situ ITS-90 scale
- standard_name :
- sea_water_temperature
- units :
- degree_Celsius
- valid_min :
- -2.5
- valid_max :
- 40.0
- C_format :
- %9.3f
- FORTRAN_format :
- F9.3
- resolution :
- 0.001
[250228 values with dtype=float32]
- TEMP_ADJUSTED_QC(N_PROF, N_LEVELS)object...
- long_name :
- quality flag
- conventions :
- Argo reference table 2
[250228 values with dtype=object]
- TEMP_ADJUSTED_ERROR(N_PROF, N_LEVELS)float32...
- long_name :
- Contains the error on the adjusted values as determined by the delayed mode QC process
- units :
- degree_Celsius
- C_format :
- %9.3f
- FORTRAN_format :
- F9.3
- resolution :
- 0.001
[250228 values with dtype=float32]
- PSAL(N_PROF, N_LEVELS)float32...
- long_name :
- Practical salinity
- standard_name :
- sea_water_salinity
- units :
- psu
- valid_min :
- 2.0
- valid_max :
- 41.0
- C_format :
- %9.3f
- FORTRAN_format :
- F9.3
- resolution :
- 0.001
[250228 values with dtype=float32]
- PSAL_QC(N_PROF, N_LEVELS)object...
- long_name :
- quality flag
- conventions :
- Argo reference table 2
[250228 values with dtype=object]
- PSAL_ADJUSTED(N_PROF, N_LEVELS)float32...
- long_name :
- Practical salinity
- standard_name :
- sea_water_salinity
- units :
- psu
- valid_min :
- 2.0
- valid_max :
- 41.0
- C_format :
- %9.3f
- FORTRAN_format :
- F9.3
- resolution :
- 0.001
[250228 values with dtype=float32]
- PSAL_ADJUSTED_QC(N_PROF, N_LEVELS)object...
- long_name :
- quality flag
- conventions :
- Argo reference table 2
[250228 values with dtype=object]
- PSAL_ADJUSTED_ERROR(N_PROF, N_LEVELS)float32...
- long_name :
- Contains the error on the adjusted values as determined by the delayed mode QC process
- units :
- psu
- C_format :
- %9.3f
- FORTRAN_format :
- F9.3
- resolution :
- 0.001
[250228 values with dtype=float32]
- PARAMETER(N_PROF, N_CALIB, N_PARAM)object...
- long_name :
- List of parameters with calibration information
- conventions :
- Argo reference table 3
array([[[b'PRES ', b'TEMP ', b'PSAL '], [b' ', b' ', b' '], [b' ', b' ', b' ']], [[b'PRES ', b'TEMP ', b'PSAL '], [b' ', b' ', b' '], [b' ', b' ', b' ']], ..., [[b'PRES ', b'TEMP ', b'PSAL '], [b' ', b' ', b' '], [b' ', b' ', b' ']], [[b'PRES ', b'TEMP ', b'PSAL '], [b' ', b' ', b' '], [b' ', b' ', b' ']]], dtype=object) - SCIENTIFIC_CALIB_EQUATION(N_PROF, N_CALIB, N_PARAM)object...
- long_name :
- Calibration equation for this parameter
array([[[b'PRES_ADJUSTED = PRES ', b'TEMP_ADJUSTED = TEMP ', b'None + dS, dS is calculated from a potential conductivity (ref to 0 dbar) multiplicative adjustment term r. '], [b' ', b' ', b' '], [b' ', b' ', b' ']], [[b'none ', b'none ', b'none '], [b' ', b' ', b' '], [b' ', b' ', b' ']], ..., [[b'PRES_ADJUSTED = PRES ', b'TEMP_ADJUSTED = TEMP ', b'PSAL_ADJUSTED = a1 * PSAL + a0 '], [b' ', b' ', b' '], [b' ', b' ', b' ']], [[b' ', b' ', b' '], [b' ', b' ', b' '], [b' ', b' ', b' ']]], dtype=object) - SCIENTIFIC_CALIB_COEFFICIENT(N_PROF, N_CALIB, N_PARAM)object...
- long_name :
- Calibration coefficients for this equation
array([[[b'None ', b'None ', b'NoneOW: r =NaN(+/-NaN), vertically averaged dS =NaN(+/-NaN), breaks: 1, Map Scales:[x=4,2; y=2,1]. '], [b' ', b' ', b' '], [b' ', b' ', b' ']], [[b' ', b' ', b' '], [b' ', b' ', b' '], [b' ', b' ', b' ']], ..., [[b'Not applicable ', b'Not applicable ', b'a1=1, a0=0.0000 '], [b' ', b' ', b' '], [b' ', b' ', b' ']], [[b' ', b' ', b' '], [b' ', b' ', b' '], [b' ', b' ', b' ']]], dtype=object) - SCIENTIFIC_CALIB_COMMENT(N_PROF, N_CALIB, N_PARAM)object...
- long_name :
- Comment applying to this parameter calibration
array([[[b'SOLO-W floats auto-correct mild pressure drift by zeroing the pressure sensor while on the surface. Additional correction was unnecessary in DMQC; PRES_ADJ_ERR: SBE sensor accuracy + resolution error ', b'No significant temperature drift detected; TEMP_ADJ_ERR: SBE sensor accuracy + resolution error ', b'No thermal lag adjustment because of insufficient data. Significant salinity drift, OW fit adopted: fit for cycles 0 to 116. The quoted error is max[0.01, 1xOW uncertainty] in PSS-78. '], [b' ', b' ', b' '], [b' ', b' ', b' ']], [[b' ', b' ', b' '], [b' ', b' ', b' '], [b' ', b' ', b' ']], ..., [[b'No adjustment performed (values duplicated) ', b'No adjustment performed (values duplicated) ', b'Real-time salinity adjustment '], [b' ', b' ', b' '], [b' ', b' ', b' ']], [[b' ', b' ', b' '], [b' ', b' ', b' '], [b' ', b' ', b' ']]], dtype=object) - SCIENTIFIC_CALIB_DATE(N_PROF, N_CALIB, N_PARAM)object...
- long_name :
- Date of calibration
- conventions :
- YYYYMMDDHHMISS
array([[[b'20210812000000', b'20210812000000', b'20210812000000'], [b' ', b' ', b' '], [b' ', b' ', b' ']], [[b' ', b' ', b' '], [b' ', b' ', b' '], [b' ', b' ', b' ']], ..., [[b'20211108080108', b'20211108080108', b'20180326000000'], [b' ', b' ', b' '], [b' ', b' ', b' ']], [[b' ', b' ', b' '], [b' ', b' ', b' '], [b' ', b' ', b' ']]], dtype=object) - HISTORY_INSTITUTION(N_HISTORY, N_PROF)object...
- long_name :
- Institution which performed action
- conventions :
- Argo reference table 4
array([], shape=(0, 188), dtype=object)
- HISTORY_STEP(N_HISTORY, N_PROF)object...
- long_name :
- Step in data processing
- conventions :
- Argo reference table 12
array([], shape=(0, 188), dtype=object)
- HISTORY_SOFTWARE(N_HISTORY, N_PROF)object...
- long_name :
- Name of software which performed action
- conventions :
- Institution dependent
array([], shape=(0, 188), dtype=object)
- HISTORY_SOFTWARE_RELEASE(N_HISTORY, N_PROF)object...
- long_name :
- Version/release of software which performed action
- conventions :
- Institution dependent
array([], shape=(0, 188), dtype=object)
- HISTORY_REFERENCE(N_HISTORY, N_PROF)object...
- long_name :
- Reference of database
- conventions :
- Institution dependent
array([], shape=(0, 188), dtype=object)
- HISTORY_DATE(N_HISTORY, N_PROF)object...
- long_name :
- Date the history record was created
- conventions :
- YYYYMMDDHHMISS
array([], shape=(0, 188), dtype=object)
- HISTORY_ACTION(N_HISTORY, N_PROF)object...
- long_name :
- Action performed on data
- conventions :
- Argo reference table 7
array([], shape=(0, 188), dtype=object)
- HISTORY_PARAMETER(N_HISTORY, N_PROF)object...
- long_name :
- Station parameter action is performed on
- conventions :
- Argo reference table 3
array([], shape=(0, 188), dtype=object)
- HISTORY_START_PRES(N_HISTORY, N_PROF)float32...
- long_name :
- Start pressure action applied on
- units :
- decibar
array([], shape=(0, 188), dtype=float32)
- HISTORY_STOP_PRES(N_HISTORY, N_PROF)float32...
- long_name :
- Stop pressure action applied on
- units :
- decibar
array([], shape=(0, 188), dtype=float32)
- HISTORY_PREVIOUS_VALUE(N_HISTORY, N_PROF)float32...
- long_name :
- Parameter/Flag previous value before action
array([], shape=(0, 188), dtype=float32)
- HISTORY_QCTEST(N_HISTORY, N_PROF)object...
- long_name :
- Documentation of tests performed, tests failed (in hex form)
- conventions :
- Write tests performed when ACTION=QCP$; tests failed when ACTION=QCF$
array([], shape=(0, 188), dtype=object)
- title :
- Argo float vertical profile
- institution :
- FR GDAC
- source :
- Argo float
- history :
- 2021-12-11T02:20:00Z creation
- references :
- http://www.argodatamgt.org/Documentation
- user_manual_version :
- 3.1
- Conventions :
- Argo-3.1 CF-1.6
- featureType :
- trajectoryProfile
Besides the core variables, TEMP, PSAL and PRES, we also have the variables TEMP_ADJUSTED, PSAL_ADJUSTED and PRES_ADJUSTED, which correspond to DM, or calibrated data. However, here we keep the focus on the Real-Time data, and in the next section, we will use the calibrated data.
Let’s begin by plotting the location of all the data.
Location#
fig, ax = plt.subplots(figsize=(20,10))
sc = ax.scatter(dayADS.LONGITUDE, dayADS.LATITUDE, cmap=qcmap)
ax.grid()
If we want to get a better map, we need cartopy, although we could skip this step.
import cartopy.crs as ccrs
import cartopy
fig,ax = plt.subplots(figsize=(20,10),subplot_kw={'projection': ccrs.PlateCarree()})
ax.plot(dayADS.LONGITUDE,dayADS.LATITUDE,'ob')
ax.add_feature(cartopy.feature.LAND)
ax.add_feature(cartopy.feature.COASTLINE, edgecolor='white')
ax.set_title(f"Data from {dayADS.JULD[0].values.astype('datetime64[D]')}")
ax.gridlines(draw_labels=True, dms=True, x_inline=False, y_inline=False)
ax.set_xlim([-100, 40]);
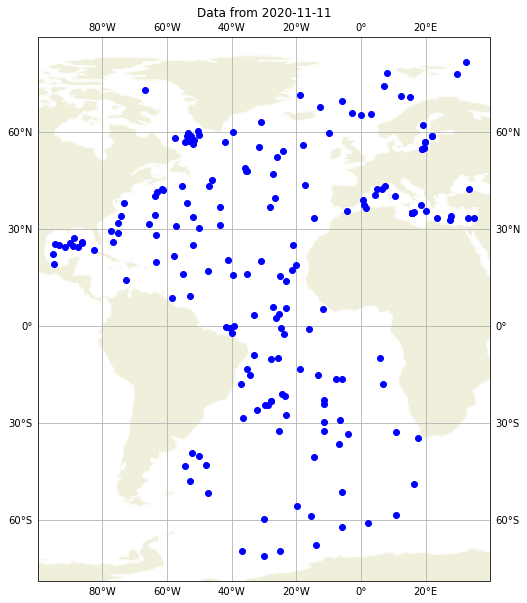
and now the actual data, a TS diagram
fig, ax = plt.subplots(figsize=(20,10))
sc = ax.scatter(dayADS.PSAL, dayADS.TEMP, cmap=qcmap)
ax.grid()
some data is obviously wrong, hence, let’s check if all the QC is Good data
pres=dayADS.PRES
lon=dayADS.LONGITUDE+pres*0
fig, ax = plt.subplots(figsize=(20,10))
sc = ax.scatter(lon, pres, c=dayADS.PSAL_QC, vmin=0, vmax=9, cmap=qcmap)
colorbar_qc(qcmap, ax=ax)
ax.grid()
ax.set_ylim(0,2000)
ax.invert_yaxis()
ax.set_xlabel(f"{dayADS.PSAL.long_name}")
ax.set_ylabel('Pressure')
ax.set_title('PSAL_QC');
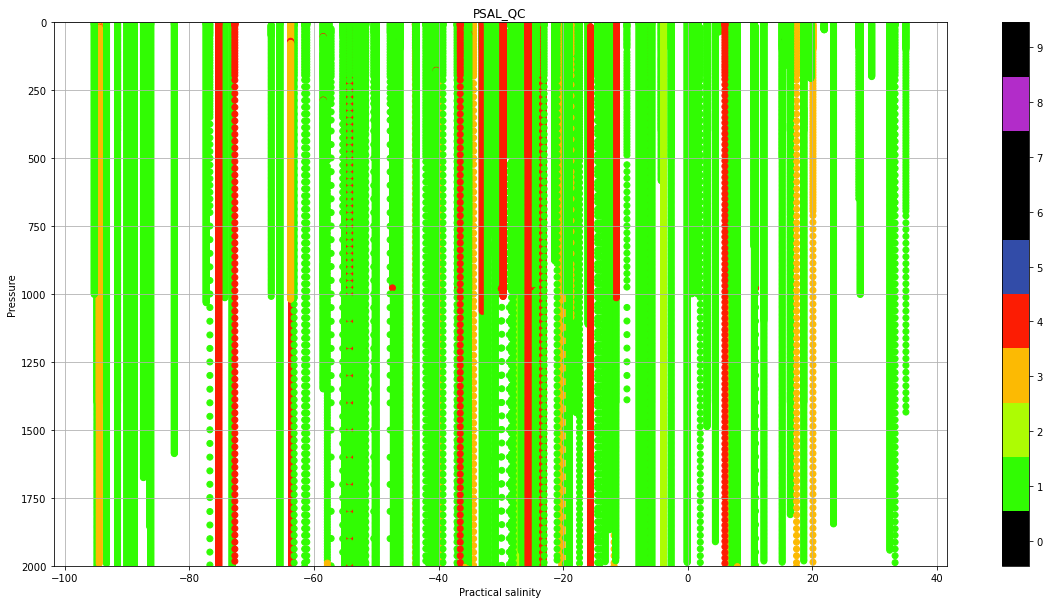
There a lot of profiles with Quality Flag (QC) that indicate bada data. Now, we will plot the same TS diagram, but color coding the data bvased on the Quality flags.
fig, ax = plt.subplots(figsize=(20,10))
sc = ax.scatter(dayADS.PSAL.where(dayADS.PSAL_QC.values.astype(float) == 0),
dayADS.TEMP.where(dayADS.PSAL_QC.values.astype(float) == 0),
c=dayADS.PSAL_QC.where(dayADS.PSAL_QC.values.astype(float) == 0), vmin=0, vmax=9, cmap=qcmap)
sc = ax.scatter(dayADS.PSAL.where(dayADS.PSAL_QC.values.astype(float) == 1),
dayADS.TEMP.where(dayADS.PSAL_QC.values.astype(float) == 1),
c=dayADS.PSAL_QC.where(dayADS.PSAL_QC.values.astype(float) == 1), vmin=0, vmax=9, cmap=qcmap)
sc = ax.scatter(dayADS.PSAL.where(dayADS.PSAL_QC.values.astype(float) == 2),
dayADS.TEMP.where(dayADS.PSAL_QC.values.astype(float) == 2),
c=dayADS.PSAL_QC.where(dayADS.PSAL_QC.values.astype(float) == 2), vmin=0, vmax=9, cmap=qcmap)
sc = ax.scatter(dayADS.PSAL.where(dayADS.PSAL_QC.values.astype(float) == 3),
dayADS.TEMP.where(dayADS.PSAL_QC.values.astype(float) == 3),
c=dayADS.PSAL_QC.where(dayADS.PSAL_QC.values.astype(float) == 3), vmin=0, vmax=9, cmap=qcmap)
sc = ax.scatter(dayADS.PSAL.where(dayADS.PSAL_QC.values.astype(float) == 4),
dayADS.TEMP.where(dayADS.PSAL_QC.values.astype(float) == 4),
c=dayADS.PSAL_QC.where(dayADS.PSAL_QC.values.astype(float) == 4), vmin=0, vmax=9, cmap=qcmap)
colorbar_qc(qcmap, ax=ax)
ax.grid()
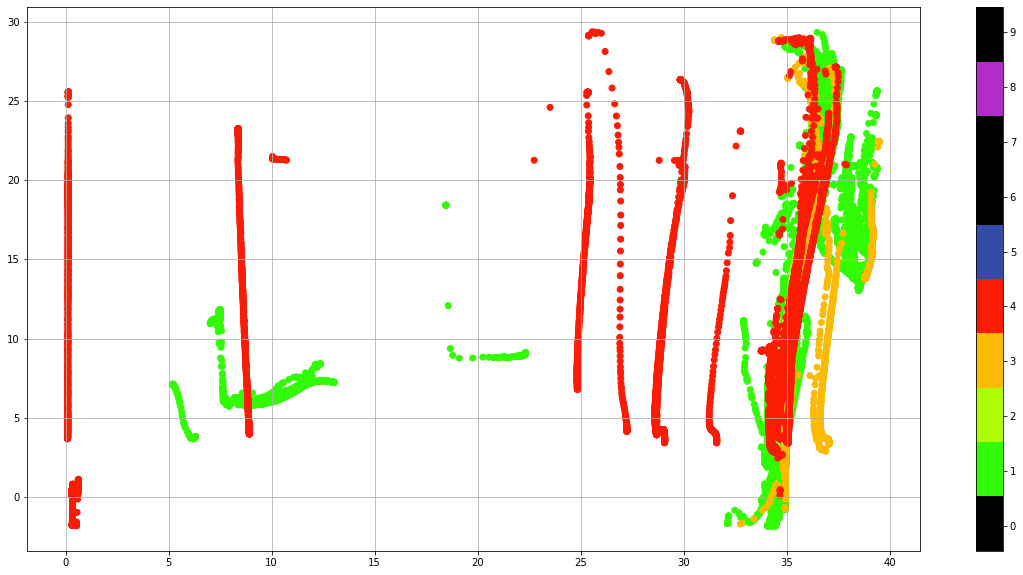
Using the QC flags we could just select the good data (QC=1) or, if we are familiar with the data, we can keep all the data that could be good, (QC=0, 1, 2 or 5) and decide what to do with the suspicius data.
Addtionally there is a Global quality flag for each one of the parameters, that indicate the percentage of good data in the profile. For salinitiy this global quality flag is PROFILE_PSAL_QC and it
Flag |
Description |
|---|---|
A |
N = 100% , all profile levels contain good data |
B |
75% <= N < 100% |
C |
75% <= N < 100% |
D |
75% <= N < 100% |
E |
75% <= N < 100% |
F |
N = 0%, no profile levels have good data |
A N = 100%; All profile levels contain good data. B 75% <= N < 100% C 50% <= N < 75% D 25% <= N < 50% E 0% < N < 25%
Example : PROFILE_TEMP_QC = A : the temperature profile contains only good values = C : the salinity profile contains 50% to 75% good values
dayADS.LONGITUDE.where(dayADS.PROFILE_PSAL_QC.values.astype(str) == 'A')
fig,ax = plt.subplots(figsize=(20,10),subplot_kw={'projection': ccrs.PlateCarree()})
ax.plot(dayADS.LONGITUDE.where(dayADS.PROFILE_PSAL_QC.values.astype(str) == 'A'),
dayADS.LATITUDE.where(dayADS.PROFILE_PSAL_QC.values.astype(str) == 'A'),
'ob')
ax.plot(dayADS.LONGITUDE.where(dayADS.PROFILE_PSAL_QC.values.astype(str) == 'B'),
dayADS.LATITUDE.where(dayADS.PROFILE_PSAL_QC.values.astype(str) == 'B'),
'or')
ax.plot(dayADS.LONGITUDE.where(dayADS.PROFILE_PSAL_QC.values.astype(str) == 'C'),
dayADS.LATITUDE.where(dayADS.PROFILE_PSAL_QC.values.astype(str) == 'D'),
'or')
ax.plot(dayADS.LONGITUDE.where(dayADS.PROFILE_PSAL_QC.values.astype(str) == 'E'),
dayADS.LATITUDE.where(dayADS.PROFILE_PSAL_QC.values.astype(str) == 'E'),
'or')
ax.plot(dayADS.LONGITUDE.where(dayADS.PROFILE_PSAL_QC.values.astype(str) == 'F'),
dayADS.LATITUDE.where(dayADS.PROFILE_PSAL_QC.values.astype(str) == 'F'),
'or')
#ax.set_title(f"Data from {Rtraj.PLATFORM_NUMBER.values.astype(str)}")
ax.add_feature(cartopy.feature.LAND)
ax.add_feature(cartopy.feature.COASTLINE, edgecolor='white')
ax.gridlines(draw_labels=True, dms=True, x_inline=False, y_inline=False)
ax.set_xlim([-100, 40]);
## QC flags for data accessed by float
iwmo = 1900379
#iwmo = 6900230
file = f"/Users/pvb/Dropbox/Oceanografia/Data/Argo/Floats/{iwmo}/{iwmo}_prof.nc"
dayADS = xr.open_dataset(file)
dayADS
<xarray.Dataset>
Dimensions: (N_PROF: 78, N_PARAM: 3, N_LEVELS: 55,
N_CALIB: 1, N_HISTORY: 0)
Dimensions without coordinates: N_PROF, N_PARAM, N_LEVELS, N_CALIB, N_HISTORY
Data variables: (12/64)
DATA_TYPE object b'Argo profile '
FORMAT_VERSION object b'3.1 '
HANDBOOK_VERSION object b'1.2 '
REFERENCE_DATE_TIME object b'19500101000000'
DATE_CREATION object b'20051214211106'
DATE_UPDATE object b'20190424095808'
... ...
HISTORY_ACTION (N_HISTORY, N_PROF) object
HISTORY_PARAMETER (N_HISTORY, N_PROF) object
HISTORY_START_PRES (N_HISTORY, N_PROF) float32
HISTORY_STOP_PRES (N_HISTORY, N_PROF) float32
HISTORY_PREVIOUS_VALUE (N_HISTORY, N_PROF) float32
HISTORY_QCTEST (N_HISTORY, N_PROF) object
Attributes:
title: Argo float vertical profile
institution: FR GDAC
source: Argo float
history: 2019-04-24T09:58:08Z creation
references: http://www.argodatamgt.org/Documentation
user_manual_version: 3.1
Conventions: Argo-3.1 CF-1.6
featureType: trajectoryProfilefig, ax = plt.subplots(figsize=(20,10))
sc = ax.scatter(dayADS.PSAL, dayADS.TEMP, cmap=qcmap)
ax.grid()
pres=dayADS.PRES
cycle=dayADS.CYCLE_NUMBER+pres*0
fig, ax = plt.subplots(figsize=(20,10))
sc = ax.scatter(cycle, pres, c=dayADS.PSAL_QC, vmin=0, vmax=9, cmap=qcmap)
colorbar_qc(qcmap, ax=ax)
ax.grid()
ax.set_ylim(0,2000)
ax.invert_yaxis()
ax.set_xlabel(f"{dayADS.PSAL.long_name}")
ax.set_ylabel('Pressure')
ax.set_title('PSAL_QC');